A Multi-center Retrospective Study [excerpt]
Journal of Digital Imaging
Joy Tzung-yu Wu, Miguel Ángel Armengol de la Hoz, Po-Chih Kuo, Joseph Alexander Paguio, Jasper Seth Yao, Edward Christopher Dee, Wesley Yeung, Jerry Jurado, Achintya Moulick, Carmelo Milazzo, Paloma Peinado, Paula Villares, Antonio Cubillo, José Felipe Varona, Hyung-Chul Lee, Alberto Estirado, José Maria Castellano & Leo Anthony Celi
Site version edited by
Joaquim Cardoso MSc.
Health Transformation Institute — for Continuous Transformation
AI Health Unit
July 5, 2022
Abstract
What is the context?
- The unprecedented global crisis brought about by the COVID-19 pandemic has sparked numerous efforts to create predictive models for the detection and prognostication of SARS-CoV-2 infections with the goal of helping health systems allocate resources.
- Machine learning models, in particular, hold promise for their ability to leverage patient clinical information and medical images for prediction.
What is the problem?
- However, most of the published COVID-19 prediction models thus far have little clinical utility due to methodological flaws and lack of appropriate validation.
What is the scope of this paper?
- In this paper, we describe our methodology to develop and validate multi-modal models for COVID-19 mortality prediction using multi-center patient data.
- The models for COVID-19 mortality prediction were developed using retrospective data from
– Madrid, Spain (N = 2547) and were externally validated in patient cohorts from a community hospital in
– New Jersey, USA (N = 242) and
– an academic center in Seoul, Republic of Korea (N = 336).
What are the results?
- The models we developed performed differently across various clinical settings, underscoring the need for a guided strategy when employing machine learning for clinical decision-making.
- We demonstrated that using features from
– both the structured electronic health records and
– chest X-ray imaging data
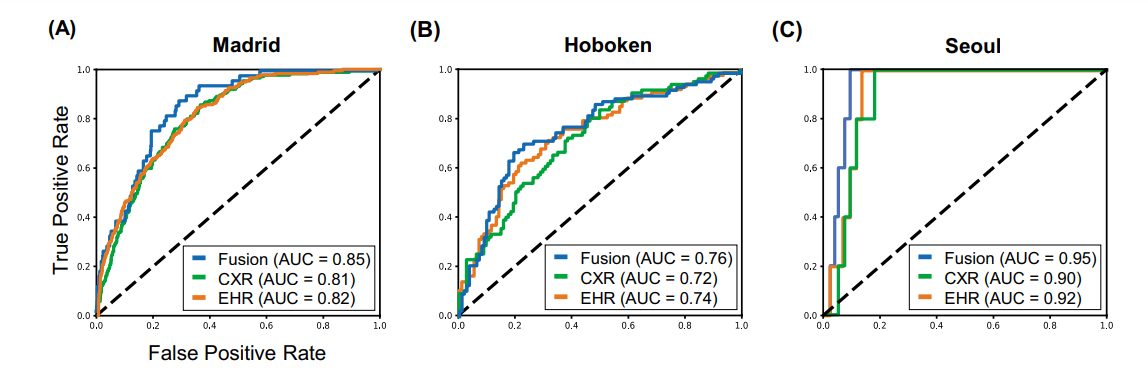
resulted in better 30-day mortality prediction performance across all three datasets (areas under the receiver operating characteristic curves: 0.85 (95% confidence interval: 0.83–0.87), 0.76 (0.70–0.82), and 0.95 (0.92–0.98)). - We discuss the rationale for the decisions made at every step in developing the models and have made our code available to the research community.
- We employed the best machine learning practices for clinical model development.
Our goal is to create a toolkit that would assist investigators and organizations in building multi-modal models for prediction, classification, and/or optimization.
The models we developed performed differently across various clinical settings, underscoring the need for a guided strategy when employing machine learning for clinical decision-making.
Selected images
Fig. 1 The proposed multi-modal models for mortality prediction
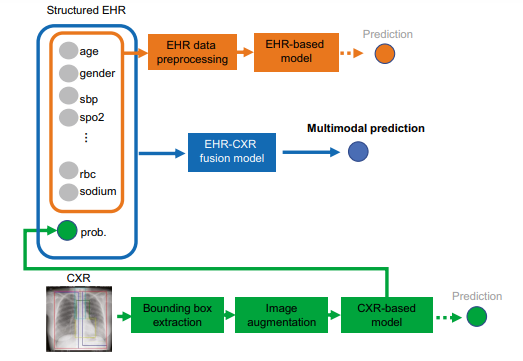
Fig. 2 A random sample of images shown to teach the model where at least 1–2 positive mortality (expired) cases are shown to the model in each batch
Fig. 3 Model performance using EHR-based model, CXR-based model, and fusion model (EHR+CXR).
(A) Internal validation on Madrid dataset; (B) external testing on Hoboken dataset; and © external testing on Seoul dataset
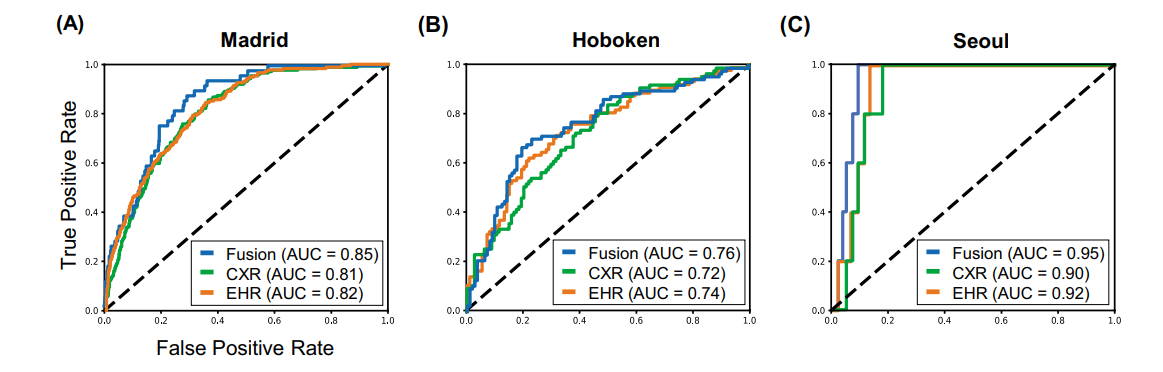
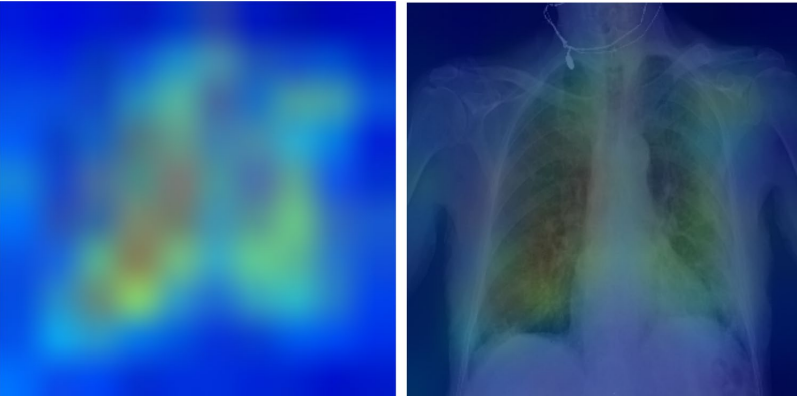
Fig. 6 Explainability: heatmaps using Grad-CAM algorithm shows that the model primarily uses imaging features from the lungs and mediastinum region for mortality prediction.
The image was produced by averaging the heatmaps from the expired patients with prediction probability larger than 0·6 and overlaying it on an actual CXR so it is easier to highlight the physiologic area
Introduction
Beginning as an outbreak of an unknown viral pneumonia in Wuhan, China, the coronavirus disease 2019 (COVID-19) pandemic has sparked numerous efforts to create predictive models.
In particular, machine learning methods hold great promise because they provide the opportunity to combine and use features from multiple modalities available in electronic health records (EHR), such as imaging and structured clinical data, for downstream prediction tasks.
At present, there are hundreds of papers in preprint servers and medical journals employing machine learning methodologies in an attempt to bridge the gaps in the diagnosis, triage, and management of COVID-19; eight of them have integrated both radiological and clinical data [ 1, 2, 3, 4, 5, 6, 7, 8].
However, most of these studies were found to have little clinical utility, producing a credibility crisis in the realm of artificial intelligence in healthcare.
A recent review by Roberts et al. found that, after screening more than 400 machine learning models using various risk and bias assessment tools, none of the evaluated machine learning models had sufficiently fulfilled all of the following:
(1) documentation of reproducible methods,
(2) adherence to best practices in the development of a model, and
(3) external validation that could justify claims of applicability [ 9].
Furthermore, the question remains as to how useful these predictive models actually are to other institutions to which these models were not customized [ 10].
While machine learning models offer the potential for a more accurate prediction of clinical outcomes within a specific context, these models were usually trained using data from a single institution and are unable to identify differences in contexts when employed in other settings [ 11].
This problem raises the need for validation not just in the neighboring center, but in other types of centers, states, or even countries, where patient demographics, standards of care, institutional policies may largely differ.
In addition, these models need to be constantly updated because the contexts in which these models were trained and approved for use may be significantly different when used at present day [ 12, 13, 14, 15].
Finally, beyond concerns about the reproducibility and generalizability of machine learning models is the issue of the lack of explainability, in which models may draw spurious associations between confounding imaging features and the outcome of interest [ 12, 16].
DeGrave et al. attempted to assess the trustworthiness of recently published machine learning models for COVID-19 by using explainable AI technology to determine which regions of chest X-rays (CXR) these models used to predict outcomes [ 12].
Surprisingly, they found that in addition to highlighting lung regions, the evaluated models used laterality marks, CXR text markers, and other features that provide no pathologic basis for distinguishing between COVID-positive and COVID-negative studies [ 12].
In other words, it was discovered that these models used shortcuts, further underscoring concerns about their applicability.
Therefore, we believe that some of the ways investigators can address the questions surrounding the credibility of a machine learning model are
(1) to state the clinical context, which include patient demographics, geography, and timeframe, of the training and testing datasets that were used
(2) to provide the resources for other centers to create or fine tune models specific to their contexts,
(3) to be explicit about the appropriate level of the model’s generalizability based on results of external validation studies,
(4) to explore strategies that either build in and/or evaluate the explainability of models, and
(5) to externally validate the performance of the model on different subpopulations of the sample and explore the fairness of the model in under-represented patient groups.
In our case, we present our efforts to develop three machine learning models for predicting 30-day mortality among hospitalized patients with COVID-19:
(1) a structured EHR-based model,
2) a CXR-based model, and
(3) an EHR-CXR fusion model.
All three models were developed using a multi-center dataset from Madrid, Spain.
We aim to investigate how the performance of each of these models differed when validated on two external unseen single-center datasets from different countries (the USA and Republic of Korea).
In addition, we will flag and detail why certain modeling design decisions were made, including the difficulties and trade-offs of these decision-making processes.
The Checklist for Artificial Intelligence in Medical Imaging [ 17] is used to report our study designs and findings.
We have made the code and other resources to reproduce our model training process available to the research community.
This work has the potential to inform triage allocation when demand exceeds hospital capacity or may aid in the prediction of the level of care, guiding inpatient assignment of patients.
We hope our work would serve as a toolkit that future investigators could use, adapt, and retrain models using data from their own institutions.
Ultimately, our goal is to provide other institutions the opportunity to leverage machine learning technology to predict the mortality of their patients with COVID-19 and customize these models to meet their individual institution’s needs.
Materials and Methods, Results and Discussion
See the original publication
Conclusion
Machining learning methods offer the advantage of utilizing richer clinical data for predictive modeling, which many have explored during the COVID-19 pandemic.
However, many studies published so far have further exposed the credibility crisis that machine learning is facing in terms of reproducibility, generalizability, explainability, and fairness.
This is often due to implementation issues, such as poorly documented study designs, lack of external test sets, and study code availability.
…many studies published so far have further exposed the credibility crisis that machine learning is facing in terms of reproducibility, generalizability, explainability, and fairness.
This is often due to implementation issues, such as poorly documented study designs, lack of external test sets, and study code availability.
In this paper, we employed best machine learning practices and trained three machine learning models on a model development (internal) dataset.
We subsequently stress tested the final models on two external datasets from different countries.
We redemonstrated (1) that using features from both the EHR and CXR imaging data resulted in better 30-day mortality prediction performances across all three datasets, and (2) the need to fine tune models on local datasets and update with time.
We evaluated our models for explainability in terms of feature dependence, and fairness in terms of gender-based performance differences.
Finally, for the sake of transparency and reproducibility, we documented all study design decisions and made the study code available to the research community.
References and Additional information
See the original publication
Cite this article
Wu, J.Ty., de la Hoz, M.Á.A., Kuo, PC. et al. Developing and Validating Multi-Modal Models for Mortality Prediction in COVID-19 Patients: a Multi-center Retrospective Study. J Digit Imaging (2022). https://doi.org/10.1007/s10278-022-00674-z
Originally published at https://link.springer.com on July 5, 2022.
About the authors & affiliations
Joy Tzung‑yu Wu1 · Miguel Ángel Armengol de la Hoz2,3,4 · Po‑Chih Kuo2,5 · Joseph Alexander Paguio6,9 · Jasper Seth Yao6,9 · Edward Christopher Dee7 · Wesley Yeung2,8 · Jerry Jurado9 · Achintya Moulick9 · Carmelo Milazzo9 · Paloma Peinado10 · Paula Villares10 · Antonio Cubillo10 · José Felipe Varona10 · Hyung‑Chul Lee11 · Alberto Estirado10 · José Maria Castellano10,12 · Leo Anthony Celi2,13,14
1 Department of Radiology and Nuclear Medicine, Stanford University, Palo Alto, CA, USA
2 Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, USA
3 Department of Anesthesia, Critical Care and Pain Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA
4 Big Data Department, Fundacion Progreso Y Salud, Regional Ministry of Health of Andalucia, Andalucia, Spain
5 Department of Computer Science, National Tsing Hua University, Hsinchu, Taiwan
6 Albert Einstein Medical Center, Philadelphia, PA, USA
7 Department of Radiation Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA
8 National University Heart Center, National University Hospital, Singapore, Singapore
9 Hoboken University Medical Center–CarePoint Health, Hoboken, NJ, USA
10 Centro Integral de Enfermedades Cardiovasculares, Hospital Universitario Monteprincipe, Grupo HM Hospitales, Madrid, Spain
11 Department of Anesthesiology and Pain Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea
12 Centro Nacional de Investigaciones Cardiovasculares, Instituto de Salud Carlos III, Madrid, Spain
13 Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA
14 Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA