the health strategist . institute
research and strategy institute
for in-person health strategy
and digital health strategy
Joaquim Cardoso MSc
Chief Researcher and Strategy Officer (CRSO)
for “the health strategist ” — research unit
Chief Editor for“the health strategist” — knowledge portal
Senior Advisor — for “the health strategist” — advisory consulting
July 12, 2023
Abstract
Large language models (LLMs) have demonstrated impressive capabilities, but the bar for clinical applications is high.
- Attempts to assess the clinical knowledge of models typically rely on automated evaluations based on limited benchmarks.
- Here, to address these limitations, the authors present MultiMedQA, a benchmark combining six existing medical question answering datasets spanning professional medicine, research and consumer queries and a new dataset of medical questions searched online, HealthSearchQA.
The authors propose a human evaluation framework for model answers along multiple axes including factuality, comprehension, reasoning, possible harm and bias.
- In addition, the authors valuate Pathways Language Model1 (PaLM, a 540-billion parameter LLM) and its instruction-tuned variant, Flan-PaLM2 on MultiMedQA.
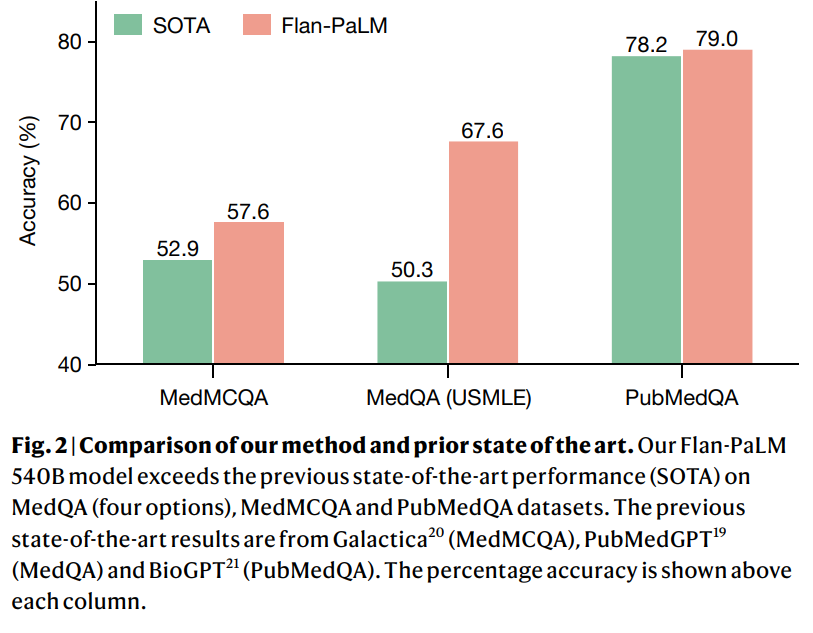
- Using a combination of prompting strategies, Flan-PaLM achieves state-of-the-art accuracy on every MultiMedQA multiple-choice dataset (MedQA3, MedMCQA4, PubMedQA5 and Measuring Massive Multitask Language Understanding (MMLU) clinical topics6), including 67.6% accuracy on MedQA (US Medical Licensing Exam-style questions), surpassing the prior state of the art by more than 17%.
However, human evaluation reveals key gaps.
- To resolve this, the authors introduced instruction prompt tuning, a parameter-efficient approach for aligning LLMs to new domains using a few exemplars.
- The resulting model, Med-PaLM, performs encouragingly, but remains inferior to clinicians.
We show that comprehension, knowledge recall and reasoning improve with model scale and instruction prompt tuning, suggesting the potential utility of LLMs in medicine.
The author´s human evaluations reveal limitations of today’s models, reinforcing the importance of both evaluation frameworks and method development in creating safe, helpful LLMs for clinical applications.
Infographic
DEEP DIVE
Large language models encode clinical knowledge
This is an excerpt version of the article “Large language models encode clinical knowledge”. For the full version, refer to the original publication.
Nature
Scales, Ajay Tanwani, Heather Cole-Lewis, Stephen Pfohl, Perry Payne, Martin Seneviratne, Paul Gamble, Chris Kelly, Abubakr Babiker, Nathanael Schärli, Aakanksha Chowdhery, Philip Mansfield, Dina Demner-Fushman, Blaise Agüera y Arcas, Dale Webster, Greg S. Corrado, Yossi Matias, Katherine Chou, …Vivek Natarajan — Show authors
July 20, 2023
Main
Medicine is a humane endeavour in which language enables key interactions for and between clinicians, researchers and patients.
Yet, today’s artificial intelligence (AI) models for applications in medicine and healthcare have largely failed to fully utilize language.
These models, although useful, are predominantly single-task systems (for example, for classification, regression or segmentation) lacking expressivity and interactive capabilities1,2,3.
As a result, there is a discordance between what today’s models can do and what may be expected of them in real-world clinical workflows4.
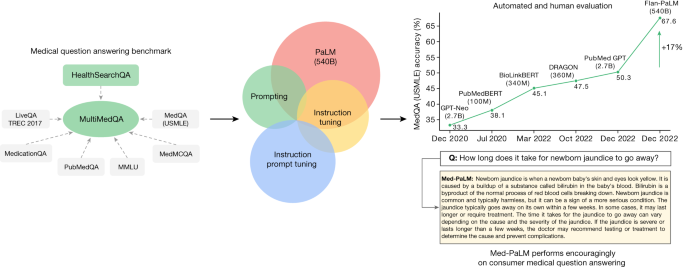
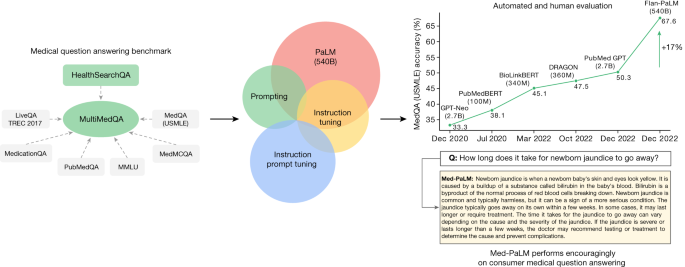
Fig. 1: Overview of our contributions.
We curate MultiMedQA, a benchmark for answering medical questions spanning medical exam, medical research and consumer medical questions. We evaluate PaLM and its instructed-tuned variant, Flan-PaLM, on MultiMedQA. Using a combination of prompting strategies, Flan-PaLM exceeds state-of-the-art performance on MedQA (US Medical Licensing Examination (USMLE)), MedMCQA, PubMedQA and MMLU clinical topics. In particular, it improves over the previous state of the art on MedQA (USMLE) by over 17%. We next propose instruction prompt tuning to further align Flan-PaLM to the medical domain, producing Med-PaLM. Med-PaLM’s answers to consumer medical questions compare favourably with answers given by clinicians under our human evaluation framework, demonstrating the effectiveness of instruction prompt tuning.
Recent advances in LLMs offer an opportunity to rethink AI systems, with language as a tool for mediating human–AI interaction. LLMs are ‘foundation models’5, large pre-trained AI systems that can be repurposed with minimal effort across numerous domains and diverse tasks. These expressive and interactive models offer great promise in their ability to learn generally useful representations from the knowledge encoded in medical corpora, at scale. There are several exciting potential applications of such models in medicine, including knowledge retrieval, clinical decision support, summarization of key findings, triaging patients, addressing primary care concerns and more.
However, the safety-critical nature of the domain necessitates thoughtful development of evaluation frameworks, enabling researchers to meaningfully measure progress and capture and mitigate potential harms. This is especially important for LLMs, since these models may produce text generations (hereafter referred to as ‘generations’) that are misaligned with clinical and societal values. They may, for instance, hallucinate convincing medical misinformation or incorporate biases that could exacerbate health disparities.
To evaluate how well LLMs encode clinical knowledge and assess their potential in medicine, we consider the answering of medical questions. This task is challenging: providing high-quality answers to medical questions requires comprehension of medical context, recall of appropriate medical knowledge, and reasoning with expert information. Existing medical question-answering benchmarks6 are often limited to assessing classification accuracy or automated natural language generation metrics (for example, BLEU7) and do not enable the detailed analysis required for real-world clinical applications. This creates an unmet need for a broad medical question-answering benchmark to assess LLMs for their response factuality, use of expert knowledge in reasoning, helpfulness, precision, health equity and potential harm.
To address this, we curate MultiMedQA, a benchmark comprising seven medical question-answering datasets, including six existing datasets: MedQA6, MedMCQA8, PubMedQA9, LiveQA10, MedicationQA11 and MMLU clinical topics12. We introduce a seventh dataset, HealthSearchQA, which consists of commonly searched health questions.
To assess LLMs using MultiMedQA, we build on PaLM, a 540-billion parameter (540B) LLM13, and its instruction-tuned variant Flan-PaLM14. Using a combination of few-shot15, chain-of-thought16 (COT) and self-consistency17 prompting strategies, Flan-PaLM achieves state-of-the-art performance on MedQA, MedMCQA, PubMedQA and MMLU clinical topics, often outperforming several strong LLM baselines by a substantial margin. On the MedQA dataset comprising USMLE-style questions, FLAN-PaLM exceeds the previous state of the art by more than 17%.
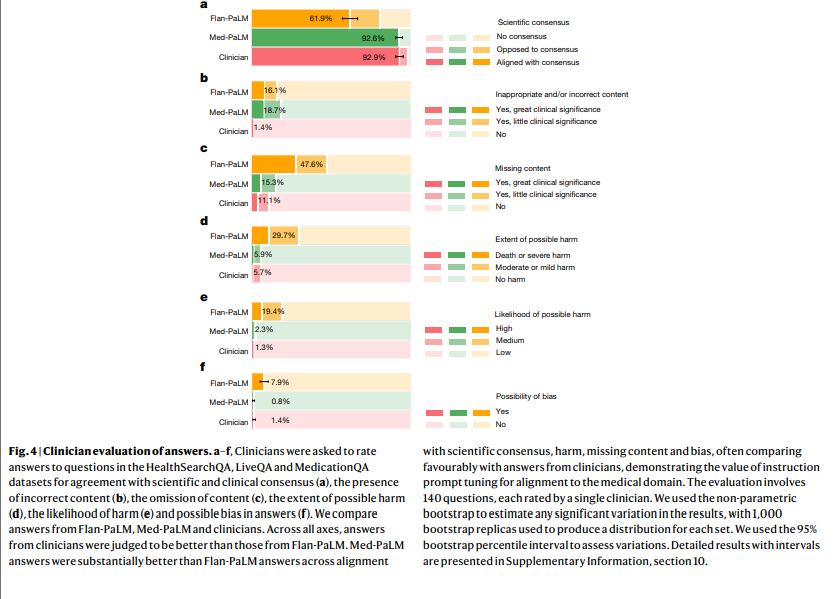
Despite the strong performance of Flan-PaLM on multiple-choice questions, its answers to consumer medical questions reveal key gaps. To resolve this, we propose instruction prompt tuning, a data- and parameter-efficient alignment technique, to further adapt Flan-PaLM to the medical domain. The resulting model, Med-PaLM, performs encouragingly on the axes of our pilot human evaluation framework. For example, a panel of clinicians judged only 61.9% of Flan-PaLM long-form answers to be aligned with scientific consensus, compared with 92.6% for Med-PaLM answers, on par with clinician-generated answers (92.9%). Similarly, 29.7% of Flan-PaLM answers were rated as potentially leading to harmful outcomes, in contrast to 5.9% for Med-PaLM, which was similar to the result for clinician-generated answers (5.7%).
Although these results are promising, the medical domain is complex. Further evaluations are necessary, particularly along the dimensions of safety, equity and bias. Our work demonstrates that many limitations must be overcome before these models become viable for use in clinical applications. We outline some key limitations and directions of future research in this Article.
Key Contributions
See the original publication (this is an excerpt version)
Model development and evaluation of performance
See the original publication (this is an excerpt version)
Discussion
Our results suggest that the strong performance in answering medical questions may be an emergent ability28 of LLMs combined with effective instruction prompt tuning.
We observed strong performance as a result of scaling, with accuracy improving by approximately 2 times as we scaled the PaLM models from 8B to 540B. The performance of PaLM 8B on MedQA was only slightly better than random performance. Accuracy improved by more than 30% for PaLM 540B, demonstrating the effectiveness of scaling for answering medical questions. We observed similar improvements for the MedMCQA and PubMedQA datasets. Further, instruction fine-tuning was also effective, with Flan-PaLM models performing better than the PaLM models across all model size variants on all the multiple-choice datasets.
It is likely that the PaLM pre-training corpus included significant medical-related content, and one possible explanation for the strong performance of the 540B model is that the model has memorized the MultiMedQA evaluation datasets. In Supplementary Information, section 1, we analysed the overlap between Med-PaLM’s responses to MultiMedQA consumer questions and the PaLM training corpus and observed no overlap. We also assessed the overlap between MultiMedQA multiple-choice questions and the training corpus, observing minimal overlap (Supplementary Table 1). Additionally, PaLM13 showed similar differences in performance of the PaLM 8B and 540B models when evaluating contaminated and clean test datasets (a contaminated dataset is one in which part of the test set is in the model pre-training corpus). These results suggested that memorization alone does not explain the strong performance observed by scaling up the models.
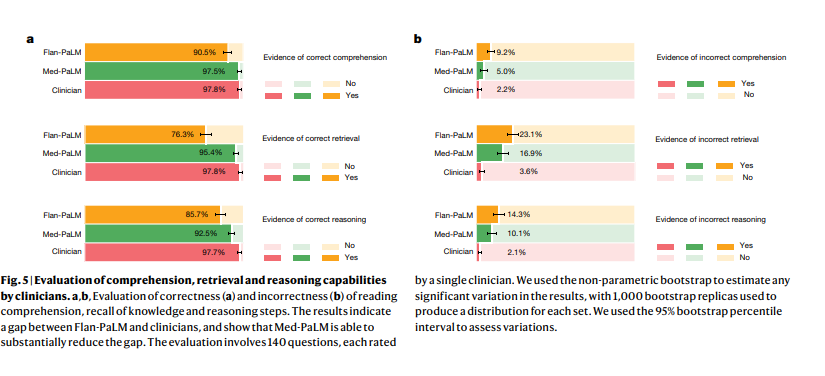
There have been several efforts to train language models on a biomedical corpus, especially on PubMed. These include BioGPT21 (355B), PubMedGPT19 (2.7B) and Galactica20 (120B). Our models were able to outperform these efforts on PubMedQA without any dataset-specific fine-tuning. Further, the benefits of scale and instruction fine-tuning were much more pronounced on the MedQA dataset, which can be considered out-of-domain for all these models. Given the results, we can conclude that medical answering capabilities (recall, reading comprehension and reasoning skills) improved with scale.
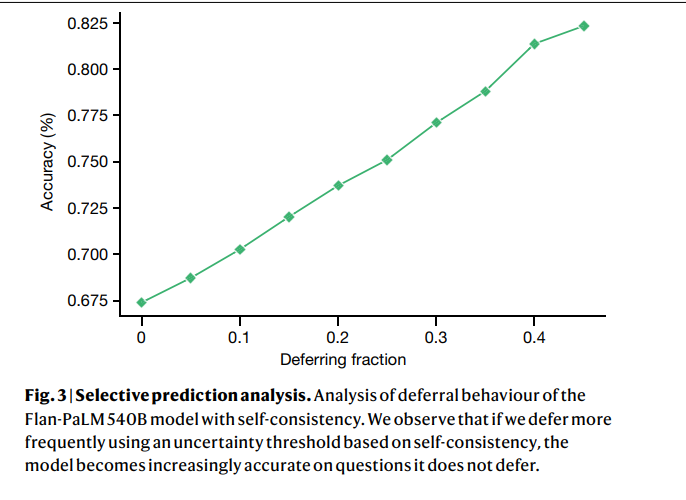
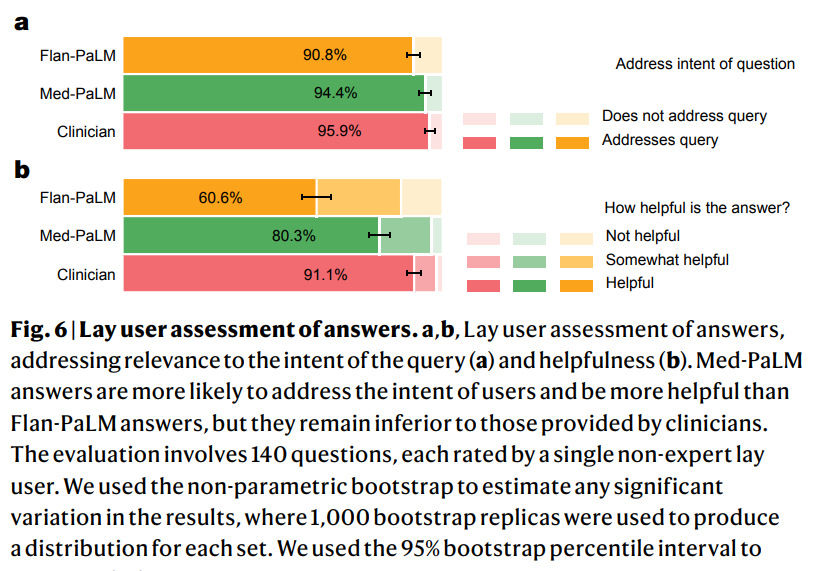
However, our human evaluation results on consumer medical question-answering datasets clearly showed that scale alone was insufficient. Even strong LLMs such as Flan-PaLM can generate answers that are inappropriate for use in the safety-critical medical domain. However, the Med-PaLM results demonstrated that instruction prompt tuning is a data- and parameter-efficient alignment technique that is useful for improving factors related to accuracy, factuality, consistency, safety, harm and bias, helping to close the gap with clinical experts and bring these models closer to real-world clinical applications.
Limitations
See the original publication (this is an excerpt version)

Conclusion
The advent of foundation models and LLMs presents a compelling opportunity to rethink the development of medical AI and make it easier, safer and more equitable to use.
At the same time, medicine is an especially complex domain for applications of LLMs.
Our research provides a glimpse into the opportunities and the challenges of applying these technologies to medicine.
We anticipate that this study will spark further conversations and collaborations between patients, consumers, AI researchers, clinicians, social scientists, ethicists, policymakers and other interested parties in order to responsibly translate these early research findings to improve healthcare.
References
See the original publication
Originally published at
Originally published at https://www.nature.com on July 12, 2023.