This is a republication of the article “A large language model for electronic health records”, published by “NPJ”, with the title above.
Institute for Continuous Health Transformation
(inHealth)
Joaquim Cardoso MSc
Founder and Chief Researcher & Advisor
December 27, 2022
Executive Summary:
What is the problem?
- There is an increasing interest in developing artificial intelligence (AI) systems to process and interpret electronic health records (EHRs).
What is the solution?
- Natural language processing (NLP) powered by pretrained language models is the key technology for medical AI systems utilizing clinical narratives.
What are the barriers? What is the evidence?
- However, there are few clinical language models, the largest of which trained in the clinical domain is comparatively small at 110 million parameters (compared with billions of parameters in the general domain).
- It is not clear how large clinical language models with billions of parameters can help medical AI systems utilize unstructured EHRs.
What is the scope of the study?
- In this study, the authors develop from scratch a large clinical language model — GatorTron — using >90 billion words of text (including >82 billion words of de-identified clinical text) and systematically evaluate it on five clinical NLP tasks including
(1) clinical concept extraction,
(2) medical relation extraction,
(3) semantic textual similarity,
(4) natural language inference (NLI), and
(5) medical question answering (MQA).
- The authors examine how (1) scaling up the number of parameters and (2) scaling up the size of the training data could benefit these NLP tasks.
What are the results?
- GatorTron models scale up the clinical language model from 110 million to 8.9 billion parameters and improve five clinical NLP tasks (e.g., 9.6% and 9.5% improvement in accuracy for NLI and MQA), …
- … which can be applied to medical AI systems to improve healthcare delivery.
GatorTron models scale up the clinical language model from 110 million to 8.9 billion parameters and improve five clinical NLP tasks
… which can be applied to medical AI systems to improve healthcare delivery.
Conclusion & recommendations
- This study demonstrates the advantages of large pretrained transformer models in the medical domain.
- GatorTron models can be applied to many other NLP tasks through fine-tuning.
- The authors believe that GatorTron will improve the use of clinical narratives in developing various medical AI systems for better healthcare delivery and health outcomes.
Infographic
ORIGINAL PUBLICATION (excerpt)
A large language model for electronic health records
npj Digital Medicine
Xi Yang, Aokun Chen, Nima PourNejatian, Hoo Chang Shin, Kaleb E. Smith, Christopher Parisien, Colin Compas, Cheryl Martin, Anthony B. Costa, Mona G. Flores, Ying Zhang, Tanja Magoc, Christopher A. Harle, Gloria Lipori, Duane A. Mitchell, William R. Hogan, Elizabeth A. Shenkman, Jiang Bian & Yonghui Wu
Introduction
There is an increasing interest in developing artificial intelligence (AI) systems to improve healthcare delivery and health outcomes using electronic health records (EHRs).
A critical step is to extract and capture patients’ characteristics from longitudinal EHRs.
The more information we have about the patients, the better the medical AI systems that we can develop.
In recent decades, hospitals and medical practices in the United States (US) have rapidly adopted EHR systems1,2, resulting in massive stores of electronic patient data, including structured (e.g., disease codes, medication codes) and unstructured (i.e., clinical narratives such as progress notes).
Even though using discrete data fields in clinical documentation has many potential advantages and structured data entry fields are increasingly added into the EHR systems, having clinicians use them remains a barrier, due to the added documentation burden3.
Physicians and other healthcare providers widely use clinical narratives as a more convenient way to document patient information ranging from family medical histories to social determinants of health4.
There is an increasing number of medical AI systems exploring the rich, more fine-grained patient information captured in clinical narratives to improve diagnostic and prognostic models5,6.
Nevertheless, free-text narratives cannot be easily used in computational models that usually require structured data.
Researchers have increasingly turned to natural language processing (NLP) as the key technology to enable medical AI systems to understand clinical language used in healthcare7.
Today, most NLP solutions are based on deep learning models8implemented using neural network architectures-a fast-developing sub-domain of machine learning.
Convolutional neural networks9(CNN) and recurrent neural networks10(RNN) have been applied to NLP in the early stage of deep learning.
More recently, the transformer architectures11(e.g., Bidirectional Encoder Representations from Transformers [BERT]) implemented with a self-attention mechanism12have become state-of-the-art, achieving the best performance on many NLP benchmarks13,14,15,16.
In the general domain, the transformer-based NLP models have achieved state-of-the-art performance for name entity recognition17,18,19, relation extraction20,21,22,23,24, sentence similarity25,26,27, natural language inference27,28,29,30, and question answering27,28,31,32.
Typically, transformers are trained in two stages: language model pretraining (i.e., learning using a self-supervised training objective on a large corpus of unlabeled text) and fine-tuning (i.e., applying the learned language models solving specific tasks with labeled training data).
One pretrained language model can be applied to solve many NLP tasks through fine-tuning, which is known as transfer learning-a strategy to learn knowledge from one task and apply it in another task33.
Human language has a very large sample space-the possible combinations of words, sentences, and their meaning and syntax are innumerable.
Recent studies show that large transformer models trained using massive text data are remarkably better than previous NLP models in terms of emergence and homogenization33.
Recent studies show that large transformer models trained using massive text data are remarkably better than previous NLP models in terms of emergence and homogenization33.
The promise of transformer models has led to further interest in exploring large-size (e.g., >billions of parameters) transformer models.
The Generative Pretrained Transformer 3 (GPT-3) model34, which has 175 billion parameters and was trained using >400 billion words of text demonstrated superior performance.
The Generative Pretrained Transformer 3 (GPT-3) model34, which has 175 billion parameters and was trained using >400 billion words of text demonstrated superior performance.
In the biomedical domain, researchers developed BioBERT11(with 110 million parameters) and PubMedBERT35(110 million parameters) transformer models using biomedical literature from PubMed.
NVIDIA developed BioMegatron models in the biomedical domain with different sizes from 345 million to 1.2 billion parameters36using a more expansive set of PubMed-derived free text.
However, few studies have explored scaling transformer models in the clinical domain due to the sensitive nature of clinical narratives that contain Protected Health Information (PHI) and the significant computing power required to increase the size of these models.
However, few studies have explored scaling transformer models in the clinical domain due to the sensitive nature of clinical narratives that contain Protected Health Information (PHI) and the significant computing power required to increase the size of these models.
To date, the largest transformer model using clinical narratives is ClinicalBERT37.
ClinicalBERT has 110 million parameters and was trained using 0.5 billion words from the publicly available Medical Information Mart for Intensive Care III38(MIMIC-III) dataset.
By developing not only larger models, but models that use clinical narratives, NLP may perform better to improve healthcare delivery and patient outcomes.
By developing not only larger models, but models that use clinical narratives, NLP may perform better to improve healthcare delivery and patient outcomes.
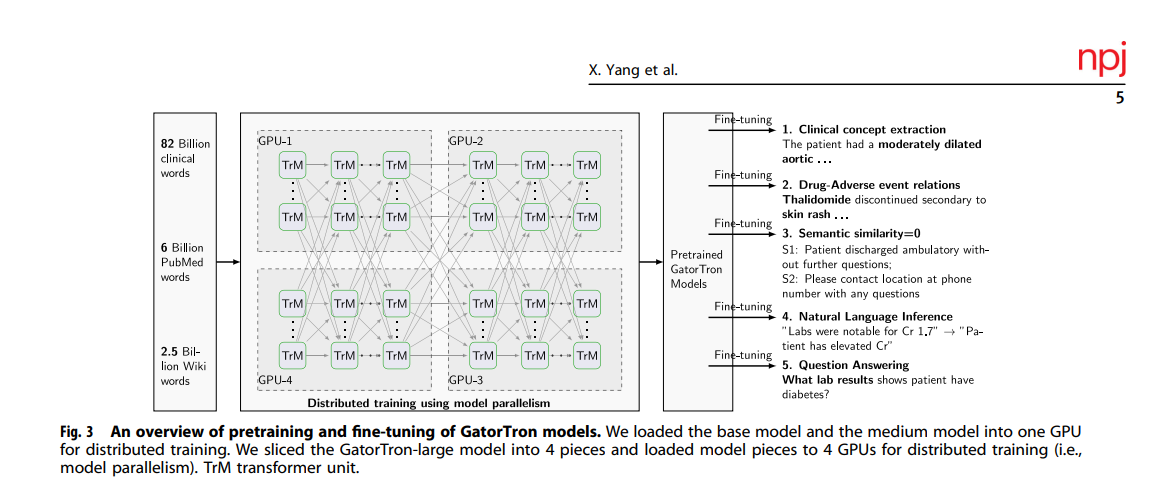
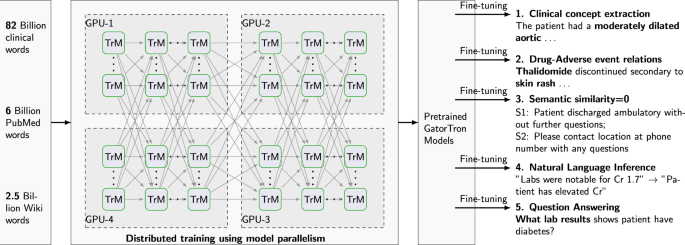
In this study, we develop a large clinical language model, GatorTron, using >90 billion words of text from the de-identified clinical notes of University of Florida (UF) Health, PubMed articles, and Wikipedia.
We train GatorTron from scratch and empirically evaluate how scaling up the number of parameters benefit the performance of downstream NLP tasks.
More specifically, we examine GatorTron models with varying number of parameters including (1) a base model with 345 million parameters, (2) a medium model with 3.9 billion parameters, and (3) a large model with 8.9 billion parameters.
We also examine how scaling up data size benefit downstream tasks by comparing the GatorTron-base model trained from the full corpus with another GatorTron-base model trained using a random sample of 1/4 of the corpus.
We compare GatorTron with existing transformer models trained using biomedical literature and clinical narratives using five clinical NLP tasks including clinical concept extraction (or named entity recognition [NER]), medical relation extraction (MRE), semantic textual similarity (STS), natural language inference (NLI), and medical question answering (MQA).
GatorTron models outperform previous transformer models from the biomedical and clinical domain on five clinical NLP tasks.
This study scales up transformer models in the clinical domain from 110 million to 8.9 billion parameters and demonstrates the benefit of large transformer models.
GatorTron models outperform previous transformer models from the biomedical and clinical domain on five clinical NLP tasks.
This study scales up transformer models in the clinical domain from 110 million to 8.9 billion parameters and demonstrates the benefit of large transformer models.

Results
A total number of 290,482,002 clinical notes from 2,476,628 patients were extracted from the UF Health Integrated Data Repository (IDR), the enterprise data warehouse of the UF Health system.
These notes were created from 2011–2021 from over 126 clinical departments and ~50 million encounters covering healthcare settings including but not limited to inpatient, outpatient, and emergency department visits.
After preprocessing and de-identification, the corpus included >82 billion medical words.
Figure 1 summarizes the distribution of patient by age, gender, race, and ethnicity as well as the distribution of notes by clinical department (top 5) and note type (top 5).
The detailed number of patients by each category, a full list of clinical departments and the corresponding proportion of notes, and a full list of note types were provided in Supplementary Table 1, Supplementary Table 2, and Supplementary Table 3.
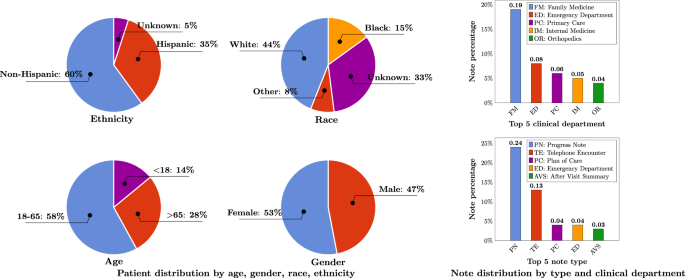
Ages were calculated as of September 2022.
Training GatorTron-large model required ~6 days on 992 A100 80 G GPUs from 124 NVIDIA DGX notes using the NVIDIA SuperPOD reference cluster architecture.
Figure 2 shows the training validation loss for all three sizes of GatorTron models.
The GatorTron-base model converged in 10 epochs, whereas the medium and large models converged in 7 epochs, which is consistent with prior observations on the faster per sample convergence of larger transformer models.
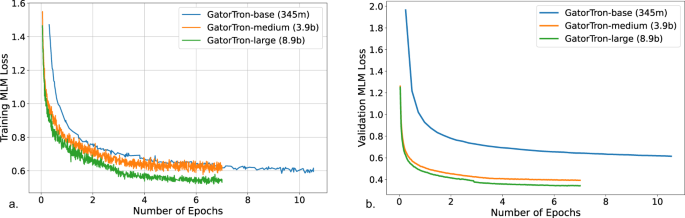
a Training loss. b Validation loss. MLM masked language modeling.
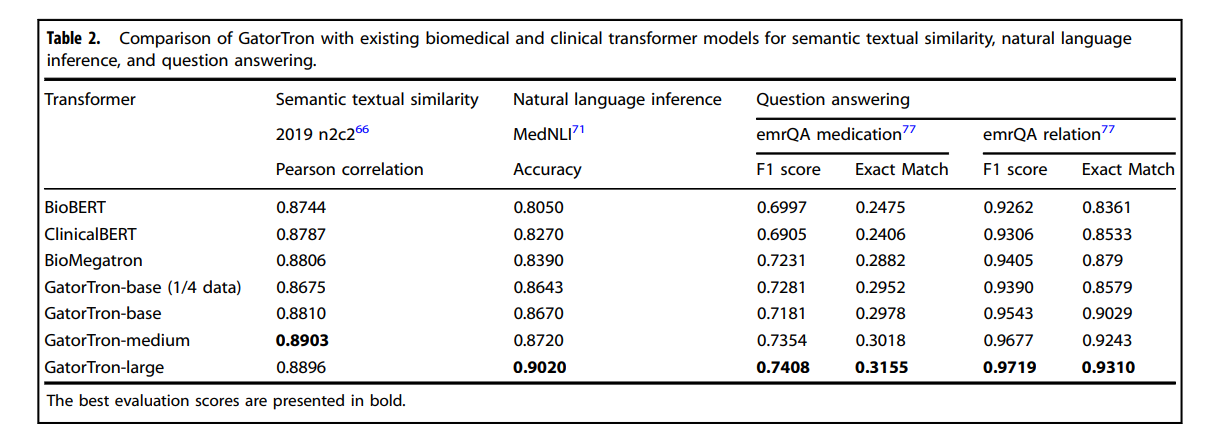
Table 1 and Table 2 compare GatorTron models with two existing biomedical transformer models (BioBERT and BioMegatron) and one clinical transformer model (Clinical BERT) on five clinical NLP tasks.
Table 1 Comparison of GatorTron with existing biomedical and clinical transformer models for clinical concept extraction and medical relation extraction.
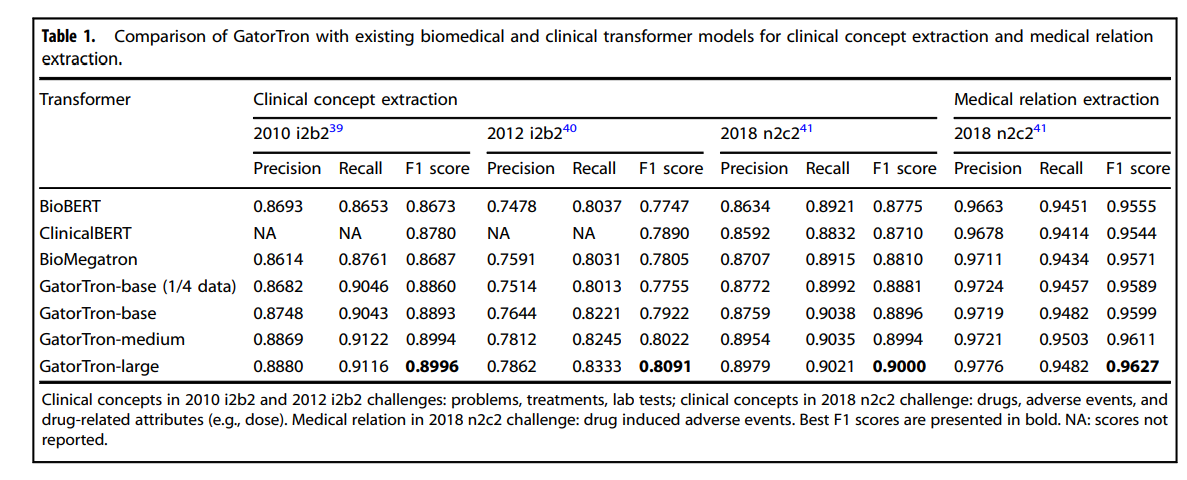
Table 2 Comparison of GatorTron with existing biomedical and clinical transformer models for semantic textual similarity, natural language inference, and question answering.
Scale up the size of training data and the number of parameters
Compared with GatorTron-base trained using a random sample of 1/4 of the corpus, the GatorTron-base model trained using the full corpus achieved improved performance for four tasks except for a sub-task in MQA (on F1 score of medication-related questions).
By scaling up the number of parameters from 345 million to 8.9 billion, GatorTron-large demonstrated remarkable improvements for all five tasks, suggesting that GatorTron models scale for canonical clinical downstream tasks and that we are not yet at the limit.
By scaling up the number of parameters from 345 million to 8.9 billion, GatorTron-large demonstrated remarkable improvements for all five tasks, suggesting that GatorTron models scale for canonical clinical downstream tasks and that we are not yet at the limit.
(1) clinical concept extraction,
(2) medical relation extraction,
(3) semantic textual similarity,
(4) natural language inference (NLI), and
(5) medical question answering (MQA).
1,2.Recognize clinical concepts and medical relations
Clinical concept extraction is to identify the concepts with important clinical meanings and classify their semantic categories (e.g., diseases, medications).
As shown in Table 1, all three GatorTron models outperformed existing biomedical and clinical transformer models in recognizing various types of clinical concepts on the three benchmark datasets (i.e., 2010 i2b239and 2012 i2b240: problem, treatments, lab tests; 2018 n2c241: drug, adverse events, and drug-related attributes).
The GatorTron-large model outperformed the other two smaller GatorTron models and achieved the best F1 scores of 0.8996, 0.8091, and 0.9000, respectively.
For medical relation extraction-a task to identify medical relations between two clinical concepts-the GatorTron-large model also achieved the best F1 score of 0.9627 for identifying drug-cause-adverse event relations outperforming existing biomedical and clinical transformers and the other two smaller GatorTron models.
We consistently observed performance improvement when scaling up the size of the GatorTron model.
3.Assess semantic textual similarity
The task of measuring semantic similarity is to determine the extent to which two sentences are similar in terms of semantic meaning.
As shown in Table 2, all GatorTron models outperformed existing biomedical and clinical transformer models.
Among the three GatorTron models, the GatorTron-medium model achieved the best Pearson correlation score of 0.8903, outperforming both GatorTron-base and GatorTron-large.
Although we did not observe consistent improvement by scaling up the size of the GatorTron model, the GatorTron-large model outperformed GatorTron-base and its performance is very close to the GatorTron-medium model (0.8896 vs. 0.8903).
4.Natural language inference
The task of NLI is to determine whether a conclusion can be inferred from a given sentence-a sentence-level NLP task.
As shown in Table 2, all GatorTron models outperformed existing biomedical and clinical transformers, and the GatorTron-large model achieved the best accuracy of 0.9020, outperforming the BioBERT and ClinicalBERT by 9.6% and 7.5%, respectively.
We observed a monotonic performance improvement by scaling up the size of the GatorTron model.
5.Medical question answering
MQA is a complex clinical NLP task that requires understand information from the entire document.
As shown in Table 2, all GatorTron models outperformed existing biomedical and clinical transformer models in answering medication and relation-related questions (e.g., “What lab results does patient have that are pertinent to diabetes diagnosis?”). For medication-related questions, the GatorTron-large model achieved the best exact match score of 0.3155, outperforming the BioBERT and ClinicalBERT by 6.8% and 7.5%, respectively. For relation-related questions, GatorTron-large also achieved the best exact match score of 0.9301, outperforming BioBERT and ClinicalBERT by 9.5% and 7.77%, respectively. We also observed a monotonic performance improvement by scaling up the size of the GatorTron model.

Discussion
In this study, we developed a large clinical transformer model, GatorTron, using a corpus of >90 billion words from UF Health (>82 billion), Pubmed (6 billion), Wikipedia (2.5 billion), and MIMIC III (0.5 billion).
We trained GatorTron with different number of parameters including 345 million, 3.9 billion, and 8.9 billion and evaluated its performance on 5 clinical NLP tasks at different linguistic levels (phrase level, sentence level, and document level) using 6 publicly available benchmark datasets. ]
The experimental results show that GatorTron models outperformed existing biomedical and clinical transformers for all five clinical NLP tasks evaluated using six different benchmark datasets.
We observed monotonic improvements by scaling up the model size of GatorTron for four of the five tasks, excluding the semantic textual similarity task.
Our GatorTron model also outperformed the BioMegatron36, a transformer model with a similar model size developed in our previous study using >8.5 billion words from PubMed and Wikipedia (a small proportion of the >90 billion words of corpus for developing GatorTron).
This study scaled up the clinical transformer models from 345 million (ClinicalBERT) to 8.9 billion parameters in the clinical domain and demonstrated remarkable performance improvements.
To the best of our knowledge, GatorTron-large is the largest transformer model in the clinical domain.
Among the five tasks, GatorTron achieved remarkable improvements for complex NLP tasks such as natural language inference and medical question answering, but moderate improvements for easier tasks such as clinical concept extraction and medical relation extraction, indicating that large transformer models are more helpful to complex NLP tasks.
These results are consistent with observations in the literature on the saturation of simpler benchmarks with large BERT architectures18,32.
GatorTron was pretrained using self-supervised masked language modeling (MLM) objective.
We monitored training loss and calculated validation loss using a subset set of the clinical text (5%) to determine the appropriate stopping time.
From the plots of training and validation losses in Fig. 2, we observed that larger GatorTron models converged faster than the smaller model.
GatorTron models perform better in extracting and interpreting patient information documented in clinical narratives, which can be integrated into medical AI systems to improve healthcare delivery and patient outcomes.
The rich, fine-grained patient information captured in clinical narratives is a critical resource powering medical AI systems.
With better performance in information extraction (e.g., clinical concept extraction and medical relation extraction), GatorTron models can provide more accurate patient information to identify research-standard patient cohorts using computable phenotypes, support physicians making data-informed decisions by clinical decision support systems, and identify adverse events associated with drug exposures for pharmacovigilance.
The observed improvements in semantic textual similarity, natural language inference, and medical question answering can be applied for deduplication of clinical text, mining medial knowledge, and developing next-generation medical AI systems that can interact with patients using human language.
We conducted error analysis and compared GatorTron with ClinicalBERT to probe the observed performance improvements.
We found that the larger, domain-specific pretrained models (e.g., GatorTron) are better at modeling longer phrases and determining semantic categories.
For example, GatorTron successfully identified “ a mildly dilated ascending aorta “, where ClinicalBERT identified only “mildly dilated” as a problem; GatorTron successfully categorized “kidney protective effects” as a “TREATMENT”, which was mis-classified as “PROBLEM” by ClinicalBERT.
For complex NLP tasks such as NLI and MQA, even large language models such as GatorTron still have difficulty in identifying the key pieces of information from longer paragraphs. Our future work will improve GatorTron in handling long pieces of text for complex NLP tasks.
This study demonstrates the advantages of large pretrained transformer models in the medical domain.
GatorTron models can be applied to many other NLP tasks through fine-tuning.
We believe that GatorTron will improve the use of clinical narratives in developing various medical AI systems for better healthcare delivery and health outcomes.
Methods & other sections
See the original publication (this is an excerpt version)
Acknowledgements
This study was partially supported by a Patient-Centered Outcomes Research Institute® (PCORI®) Award (ME-2018C3–14754), a grant from the National Cancer Institute, 1R01CA246418 R01, grants from the National Institute on Aging, NIA R56AG069880 and R21AG062884, and the Cancer Informatics and eHealth core jointly supported by the UF Health Cancer Center and the UF Clinical and Translational Science Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions. We would like to thank the UF Research Computing team, led by Dr. Erik Deumens, for providing computing power through UF HiPerGator-AI cluster.
References and additional information
See the original publication
Originally published at https://www.nature.com on December 26, 2022.
Cite this article
Yang, X., Chen, A., PourNejatian, N. et al. A large language model for electronic health records. npj Digit. Med. 5, 194 (2022).
About the authors & affiliations
Xi Yang1,2, Aokun Chen1,2, Nima PourNejatian3 , Hoo Chang Shin3 , Kaleb E. Smith3 , Christopher Parisien3 , Colin Compas3 , Cheryl Martin3 , Anthony B. Costa3 , Mona G. Flores 3 , Ying Zhang 4 , Tanja Magoc5 , Christopher A. Harle1,5, Gloria Lipori5,6, Duane A. Mitchell6 , William R. Hogan 1 , Elizabeth A. Shenkman 1 , Jiang Bian 1,2 and Yonghui Wu 1,2
1 Department of Health Outcomes and Biomedical Informatics, College of Medicine, University of Florida, Gainesville, FL, USA.
2 Cancer Informatics and eHealth core, University of Florida Health Cancer Center, Gainesville, FL, USA.
3 NVIDIA, Santa Clara, CA, USA.
4 Research Computing, University of Florida, Gainesville, FL, USA.
5 Integrated Data Repository Research Services, University of Florida, Gainesville, FL, USA.
6 Lillian S. Wells Department of Neurosurgery, UF Clinical and Translational Science Institute, University of Florida, Gainesville, FL, USA
GatorTron models are publicly available at: https://catalog.ngc.nvidia.com/orgs/nvidia/teams/clara/models/gatortron_og.