This is a republication of the article “Study: New Wastewater Surveillance Method Detected SARS-CoV-2 Variants of Concern Up to 2 Weeks Before Clinical Tests”, with the title above.
JAMA Network
Jocelyn Solis-Moreira
August 24, 2022 (iStock.com/Chunyip Wong)
Executive Summary by:
Joaquim Cardoso MSc.
health transformation — journal
September 18, 2022
- Most people with current or recent SARS-CoV-2 infections shed the virus RNA in their stool, regardless of whether they’re symptomatic, and fragments of this RNA are detectable in wastewater samples.
- Monitoring wastewater provides an efficient snapshot of the types and amounts of viruses and bacteria spreading in a community, which can augment data from clinical tests.
- A single wastewater sample gives a substantial amount of information about an entire building, town, or county, making it cost-effective.
- “[Y]ou would have to sequence a ton of clinical or nasal swabs to get that level of resolution,” .
- The new study suggests that, in addition to tracking SARS-CoV-2 transmission levels, wastewater surveillance also could be used to detect emerging viral variants when they first appear in an area.
SELECTED IMAGES
Fig. 1: Campus sampling locations and SARS-CoV-2 testing statistics.

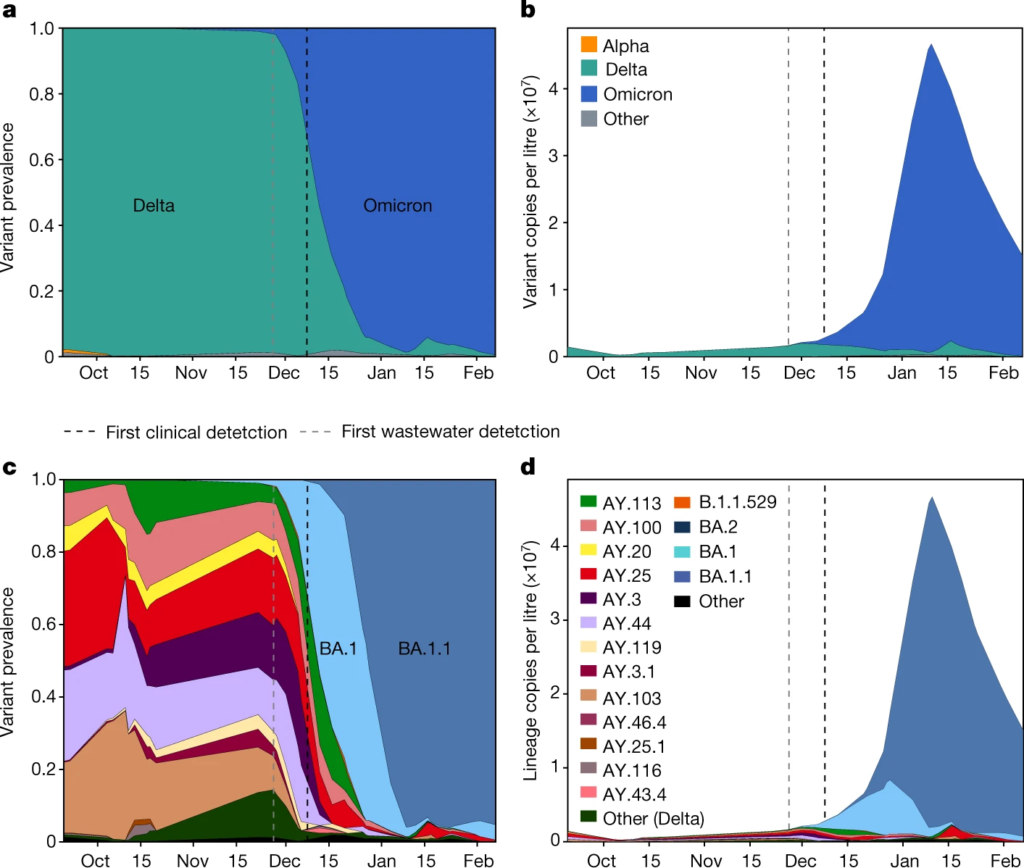
Fig. 5: Community wastewater enables early Omicron detection and reveals lineage dynamics.
ORIGINAL PUBLICATION (full version)

Researchers used a recently developed method of wastewater genomic surveillance to detect SARS-CoV-2 infections on the University of California, San Diego (UCSD), campus during 10 months of the pandemic.
The approach identified viral variants of concern as early as 2 weeks before the variants showed up in clinical tests, the team recently reported in Nature.

The Backstory
At least 55 countries already use wastewater surveillance to track SARS-CoV-2. In the US, the Centers for Disease Control and Prevention (CDC) launched a National Wastewater Surveillance System in September 2020.
The program works with health departments across the country to monitor SARS-CoV-2 levels in community wastewater.
However, detecting and identifying multiple SARS-CoV-2 variants mixed in sewage waste remains technically and economically challenging, especially on a large scale.
In the recent study, UCSD researchers and colleagues at the CDC, the California Department of Public Health, and the California Health and Human Services Agency, among other collaborators, optimized the technique and created an efficient tracking system with community results available in just a few hours.

The Technique
Nanomagnetic beads with high affinity for viral particles were added to raw sewage samples, where they captured SARS-CoV-2 and other respiratory viruses.
Robots with a magnetic head extracted the virus-bound beads in less than an hour.
The researchers then used polymerase chain reaction, or PCR, testing to decide which samples to sequence.
Karthikeyan said the magnetic nanobeads helped retrieve greater proportions of unfragmented SARS-CoV-2 RNA than other wastewater sampling methods, which makes the technique more amenable to high-quality sequencing even at low viral loads.
For example, the method allowed the team to sequence almost 95% of the virus genome, even for wastewater samples with low amounts of virus, whereas other wastewater collection techniques in development have sequenced 40% or less of the genome.
The next advance was computational.
The scientists developed an algorithm called Freyja to better identify multiple variants in a virally diverse wastewater sample and estimate how these variants are related to other SARS-CoV-2 lineage or sublineages also found in wastewater.
Freyja uses a library of “barcodes” that contain information on the specific mutations in each of the more than 1000 SARS-CoV-2 lineages.
Researchers can run the Freyja algorithm to map the shreds of viral RNA in wastewater samples to the corresponding lineages.

The Design
The researchers chose to compare clinical and wastewater genomic surveillance on the UCSD campus because places of communal living are ideal for viral spread and are also well-controlled and relatively isolated settings.
They eventually expanded their study to the greater San Diego area. The resulting 21 383 wastewater samples included:
- 19 944 samples collected from November 2020 through September 2021 from the sewage pipes of 360 UCSD campus buildings
- 1475 samples collected from September 2021 through February 2022 from the Point Loma wastewater treatment plant serving 2.3 million people and 17 public schools making up 4 school districts
UCSD students were notified by email if their residential building’s sewage sample tested positive for SARS-CoV-2.
A university dashboard also was updated every evening with the latest wastewater sampling data.
This included an interactive map that allowed students to see in real-time which buildings had positive results for SARS-CoV-2.
Staff and students at the county and public schools were offered on-site testing when a positive signal was detected.
The researchers also compared SARS-CoV-2 genome sequences from wastewater surveillance and clinical tests:
- On campus, 600 genomes from wastewater samples were compared with 759 genomes from nasal swabs.
- In the greater San Diego area, 837 genomes from county wastewater samples, including those from the UCSD campus, were compared with 31 149 genomes from nasal swabs.

What We’ve Learned
The new approach detected:
- The Alpha and Delta variants 2 weeks before nasal swab tests confirmed their presence in San Diego
- The Omicron variant 10 days before the county’s first positive clinical test
- When BA.1 Omicron variant infections overtook Delta infections in late 2021
- SARS-CoV-2 transmission that was missed by clinical genomic surveillance
In an interview, Natalie Exum, PhD, an assistant scientist in the Environmental Health and Engineering department at the Johns Hopkins Bloomberg School of Public Health who was not affiliated with the study, said the findings strongly support using sewage waste surveillance as an early detection system for new variants.
“It’s challenging because these viruses have been ripped apart, and you now have to piece them back together again,” she said of wastewater genomic surveillance.
“But what makes this paper so important is that it’s documenting that it is possible and could short-circuit the long process of getting tested and reporting [clinical] results to the CDC.”
“But what makes this paper so important is that it’s documenting that it is possible and could short-circuit the long process of getting tested and reporting [clinical] results to the CDC.”

The Barriers
Exum said starting up or expanding wastewater genomic surveillance programs, including this new approach, will initially incur an enormous cost to train people to operate the equipment.
In the long run, however, the new approach could help detect and measure SARS-CoV-2 variants in a community quicker and more cheaply than clinical swab testing alone, she explained.
In the long run, however, the new approach could help detect and measure SARS-CoV-2 variants in a community quicker and more cheaply than clinical swab testing alone, she explained.
Another concern is that it’s unclear how useful wastewater data will be in curbing viral spread.
Exum pointed out that the CDC has made national wastewater data available to the public but that the efforts are futile if individuals don’t use the information.
“I think people are just tired and have made risk-based decisions that mild coronavirus is not enough of a concern to put masks on and take preventative measures,” she said.
“There’s probably about 10% of the population that would actually decide to [do so] based on this data.”

Looking Ahead
Karthikeyan and her team have begun using their wastewater genomic surveillance program to track the monkeypox virus on the UCSD campus and in San Diego county.
They are also expanding the program to sequence other viruses and pathogens in community wastewater.
Among the pathogens they might find are viruses thought to be eradicated.
The UK’s wastewater surveillance program identified poliovirus in London’s sewage in late June of this year.
The UK’s wastewater surveillance program identified poliovirus in London’s sewage in late June of this year.
This July, the US reported its first case of community transmission since 2005 in upstate New York’s Rockland County.
In that case, vaccine-derived poliovirus was detected in the stool samples of an unvaccinated patient hospitalized with flaccid lower limb weakness.
Health officials have since detected the virus in stored wastewater samples from Rockland County and neighboring Orange County dating back to this April.
And on August 12, officials announced that the virus had been detected in New York City wastewater.
The findings suggest there may be yet another viral outbreak underway, further emphasizing the need for better surveillance of pathogens both old and new.
The findings suggest there may be yet another viral outbreak underway, further emphasizing the need for better surveillance of pathogens both old and new.
References and additional information:
See the original publication
Originally published at: https://jamanetwork.com
Names mentioned
Smruthi Karthikeyan, PhD, MS, a UCSD postdoctoral scholar and lead author of the recent study, said in an interview with JAMA.
REFERENCE PUBLICATION (reference)

Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission
Smruthi Karthikeyan, Joshua I. Levy, Peter De Hoff, Greg Humphrey, Amanda Birmingham, Kristen Jepsen, Sawyer Farmer, Helena M. Tubb, Tommy Valles, Caitlin E. Tribelhorn, Rebecca Tsai, Stefan Aigner, Shashank Sathe, Niema Moshiri, Benjamin Henson, Adam M. Mark, Abbas Hakim, Nathan A. Baer, Tom Barber, Pedro Belda-Ferre, Marisol Chacón, Willi Cheung, Evelyn S. Cresini, Emily R. Eisner, …Rob Knight et all