The Lancet — Digital Health
Mohammad Ilyas
NOVEMBER 01, 2022
The Article by Kexin Ding and colleaguesb1 uses artificial intelligence (AI) to infer somatic molecular changes (of both genome and proteome) from digital images of colorectal cancers.
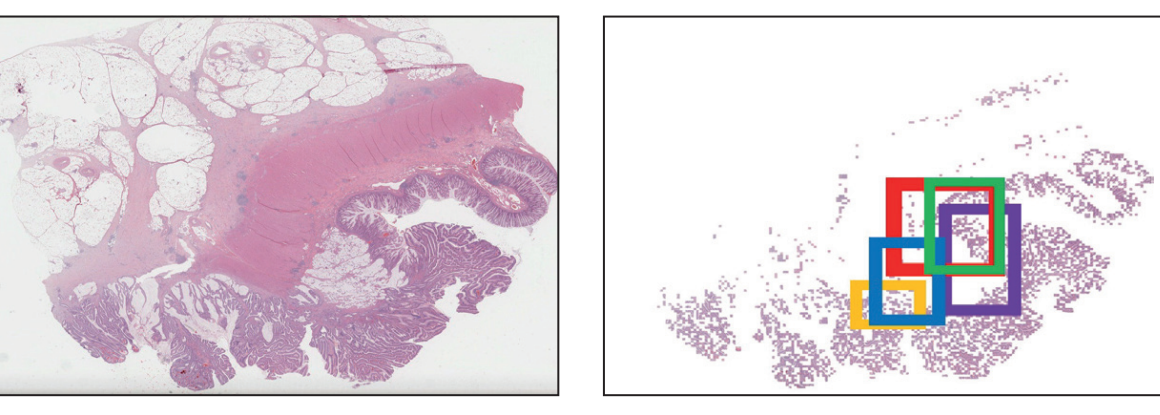
The authors made use of AI to initially segment (ie, separate out) tumour areas and then interrogate small areas (called tiles) within the tumour.
The paper extends beyond other studies by interrogating the tiles and their spatial connections using a graph neural network (GNN).
Since the tiles contain both cancer cells and non-malignant stromal cells (which comprises the tumour microenvironment), the GNN allows tumour heterogeneity to be analysed.
Using publicly available datasets, Ding and colleagues reached a similar level of accuracy to other studies that have used the same datasets but different algorithms.
The consistent results between differing studies affirms the notion that molecular data can be extracted from digital images.
Analysis can start immediately on images becoming available, which is attractive as, theoretically, it will allow a workflow whereby histological and computational analyses are completed simultaneously.
Using publicly available datasets, Ding and colleagues reached a similar level of accuracy to other studies that have used the same datasets but different algorithms.
The consistent results between differing studies affirms the notion that molecular data can be extracted from digital images.

However, biomedical research history is replete with failed biomarker studies and, although AI in cancer pathology is in its infancy, several obstacles are visible.
Firstly, real-world data, in contrast to the curated images used in research, will contain a lot more noise.
Tissues are fixed in formalin before sections are cut and variations in the fixation time can affect the molecular profile 7 -under-fixation might lead to tissue autolysis while over-fixation might alter the chemical properties of nucleic acids and proteins.
In a large resection specimen, there will be variability in fixation; around five blocks of tumour are taken for colorectal cancers and currently there are no criteria for block selection nor data on how many blocks should be analysed.
Most studies perform a pre-processing step on images to normalise the staining and exclude images that are out of focus.
However, technical artifacts can result in tissue folds, heterogeneity in stain uptake (even within small tissue fragments), or small patches within an image being out of focus.
In addition, where neoadjuvant therapy is being considered, decisions need to be made on biopsy specimens.
These specimens might contain only small numbers of malignant cells-sufficient for diagnosis but possibly problematic for image analysis.

The most important consideration in the integration of AI into the clinical care pathway is accuracy.
The highest AUC for gene mutation detection in the Article was 87·08 (95% CI 83·28–90·82).
Although this result is excellent for research, clinicians might not deem this sufficiently accurate when deciding to administer or withhold chemotherapies.
Even if algorithms become as accurate as genomic technologies (such as next-generation sequencing, NGS), it is unlikely that genomics will be replaced in the near future.
Even if algorithms become as accurate as genomic technologies (such as next-generation sequencing, NGS), it is unlikely that genomics will be replaced in the near future.
Genomic technologies are becoming cheaper and quicker.
Additionally, although AI algorithms can only inform on the biomarkers for which they have been trained, genomic technologies can provide pharmacogenomic data and inform on the generation of potentially targetable neoantigens.
Although NGS can quantify mutations and inform on mutation-carrying tumour subclones, it is uncertain how AI algorithms will handle molecular heterogeneity where only a proportion of tumour tiles indicate actionable mutation.
If both AI image analysis and genomic analysis are made use of for a tumour, a clinical (and potentially legal) quandary would arise if one form of analysis identified an actionable mutation and the other did not.
In these cases, clinical decisions would probably be made on the genomic data.
If both AI image analysis and genomic analysis are made use of for a tumour, a clinical (and potentially legal) quandary would arise if one form of analysis identified an actionable mutation and the other did not. In these cases, clinical decisions would probably be made on the genomic data.

What is the role of AI in cancer pathology if genomic data are considered superior (in the immediate future at least)?
Computational image analysis can use pixel-level, object-level, and higher semantic-level data and can therefore extract huge amounts of information.
The paucity of annotated images for use in training has hindered progress in AI algorithm development.
However, weakly supervised algorithms can obviate the need for annotation, although they require huge amounts of data.
Diagnostic histopathology is undergoing digital transformation, which means that eventually every single tissue section will be scanned, and the resulting images used for primary diagnosis.
Availability of data will therefore no longer be a limiting factor, which will enable AI algorithms to be developed for decision support in primary diagnosis (for both cancer and non-cancer diseases).
Data availability will also allow digital algorithms to be trained on a variety of clinical features such as prognosis, cancer recurrence or metastasis, and responsiveness to therapy.
In combination with genomic and other data, algorithms (such as those used by Ding and colleagues) could provide more precise information for patient management.
In combination with genomic and other data, algorithms (such as those used by Ding and colleagues) could provide more precise information for patient management.
The answer to the question in the title is, inevitably, that AI in cancer pathology has a lot of hope but we should be wary of the hype.
The answer to the question in the title is, inevitably, that AI in cancer pathology has a lot of hope but we should be wary of the hype.
References and Additional Information
See the original publication
About the authors & affiliations
Mohammad Ilyas
Unit of Translational Medical Sciences, Department of Pathology, , University of Nottingham, Queen’s Medical Centre, Nottingham, NG7 2UH, UK
Originally published at https://www.thelancet.com.
RELATED PAPER

Spatially aware graph neural networks and cross-level molecular profile prediction in colon cancer histopathology: a retrospective multi-cohort study
Kexin Ding, MS *
Mu Zhou, PhD *
He Wang, MD
Shaoting Zhang, PhD †
Prof Dimitri N Metaxas, PhD
Summary
Background
Digital whole-slide images are a unique way to assess the spatial context of the cancer microenvironment. Exploring these spatial characteristics will enable us to better identify cross-level molecular markers that could deepen our understanding of cancer biology and related patient outcomes.
Methods
We proposed a graph neural network approach that emphasises spatialisation of tumour tiles towards a comprehensive evaluation of predicting cross-level molecular profiles of genetic mutations, copy number alterations, and functional protein expressions from whole-slide images. We introduced a transformation strategy that converts whole-slide image scans into graph-structured data to address the spatial heterogeneity of colon cancer. We developed and assessed the performance of the model on The Cancer Genome Atlas colon adenocarcinoma (TCGA-COAD) and validated it on two external datasets (ie, The Cancer Genome Atlas rectum adenocarcinoma [TCGA-READ] and Clinical Proteomic Tumor Analysis Consortium colon adenocarcinoma [CPTAC-COAD]). We also predicted microsatellite instability and result interpretability.
Findings
The model was developed on 459 colon tumour whole-slide images from TCGA-COAD, and externally validated on 165 rectum tumour whole-slide images from TCGA-READ and 161 colon tumour whole-slide images from CPTAC-COAD. For TCGA cohorts, our method accurately predicted the molecular classes of the gene mutations (area under the curve [AUCs] from 82·54 [95% CI 77·41–87·14] to 87·08 [83·28–90·82] on TCGA-COAD, and AUCs from 70·46 [61·37–79·61] to 81·80 [72·20–89·70] on TCGA-READ), along with genes with copy number alterations (AUCs from 81·98 [73·34–89·68] to 90·55 [86·02–94·89] on TCGA-COAD, and AUCs from 62·05 [48·94–73·46] to 76·48 [64·78–86·71] on TCGA-READ), microsatellite instability (MSI) status classification (AUC 83·92 [77·41–87·59] on TCGA-COAD, and AUC 61·28 [53·28–67·93] on TCGA-READ), and protein expressions (AUCs from 85·57 [81·16–89·44] to 89·64 [86·29–93·19] on TCGA-COAD, and AUCs from 51·77 [42·53–61·83] to 59·79 [50·79–68·57] on TCGA-READ). For the CPTAC-COAD cohort, our model predicted a panel of gene mutations with AUC values from 63·74 (95% CI 52·92–75·37) to 82·90 (73·69–90·71), genes with copy number alterations with AUC values from 62·39 (51·37–73·76) to 86·08 (79·67–91·74), and MSI status prediction with AUC value of 73·15 (63·21–83·13).
Interpretation
We showed that spatially connected graph models enable molecular profile predictions in colon cancer and are generalised to rectum cancer. After further validation, our method could be used to infer the prognostic value of multiscale molecular biomarkers and identify targeted therapies for patients with colon cancer.
Funding
This research has been partially funded by ARO MURI 805491, NSF IIS-1793883, NSF CNS-1747778, NSF IIS 1763523, DOD-ARO ACC-W911NF, and NSF OIA-2040638 to Dimitri N Metaxas.
Originally published at https://www.thelancet.com