institute for health transformation (inhealth)
Joaquim Cardoso MSc — Founder and CEO
January 19, 2023
KEY MESSAGES
- The authors show that local ML models (Machine Learning) relying on whole-slide images can predict response to NACT …
- … but that collaborative training of ML models further improves performance, on par with the best current approaches in which ML models are trained using time-consuming expert annotations.
- This proof of concept study, in which federated learning is applied to real-world datasets, paves the way for future biomarker discovery using unprecedentedly large datasets.
INFOGRAPHIC

Nature Medicine
Abstract
Triple-negative breast cancer (TNBC) is a rare cancer, characterized by high metastatic potential and poor prognosis, and has limited treatment options.
- The current standard of care in nonmetastatic settings is neoadjuvant chemotherapy (NACT), but treatment efficacy varies substantially across patients.
- This heterogeneity is still poorly understood, partly due to the paucity of curated TNBC data.
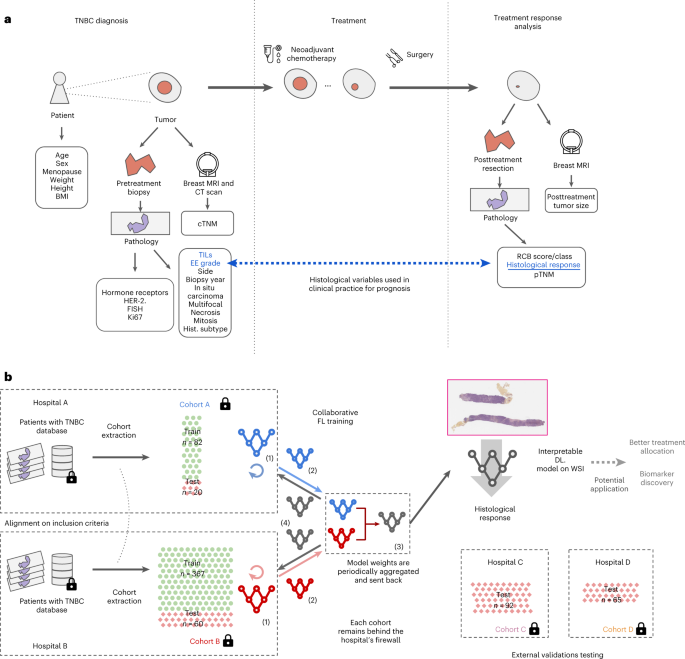
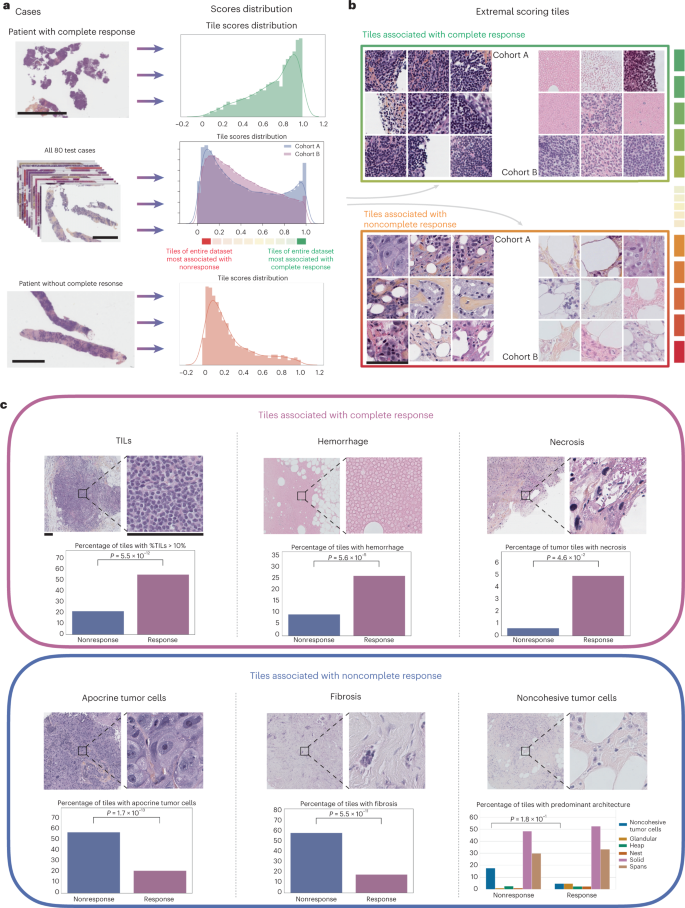
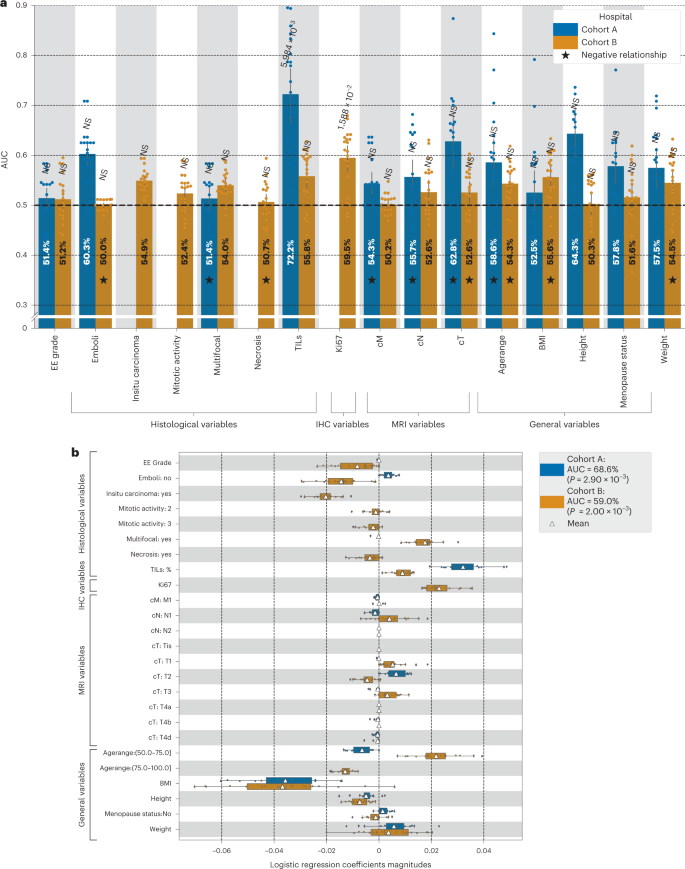
Here we investigate the use of machine learning (ML) leveraging whole-slide images and clinical information to predict, at diagnosis, the histological response to NACT for early TNBC women patients.
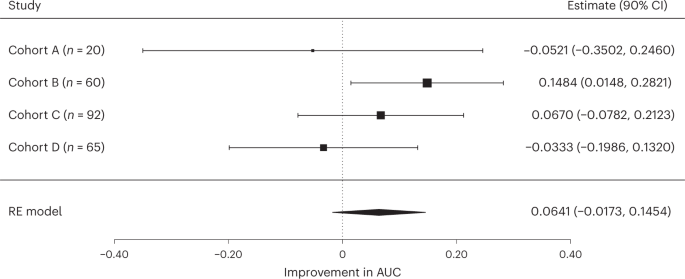
- To overcome the biases of small-scale studies while respecting data privacy, we conducted a multicentric TNBC study using federated learning, in which patient data remain secured behind hospitals’ firewalls.
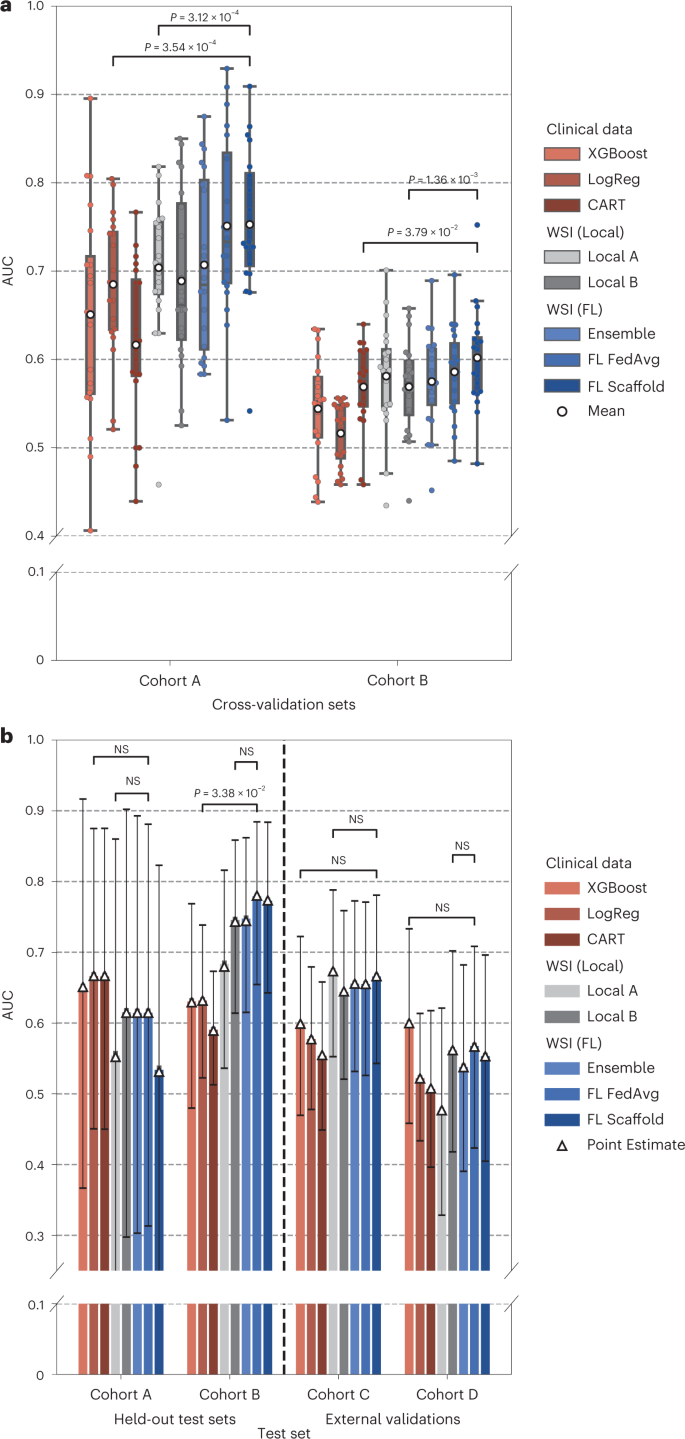
- We show that local ML models relying on whole-slide images can predict response to NACT but that collaborative training of ML models further improves performance, …
- … on par with the best current approaches in which ML models are trained using time-consuming expert annotations.
- Our ML model is interpretable and is sensitive to specific histological patterns.
This proof of concept study, in which federated learning is applied to real-world datasets, paves the way for future biomarker discovery using unprecedentedly large datasets.
Data availability
Authors and Affiliations
Owkin, Inc., New York, NY, USA
Jean Ogier du Terrail, Constance Béguier, Mathieu Andreux, Charles Maussion, Benoît Schmauch, Eric W. Tramel, Etienne Bendjebbar, Mikhail Zaslavskiy, Gilles Wainrib, Kelvin Moutet, Clément Gautier, Inal Djafar, Anne-Laure Moisson, Camille Marini, Mathieu Galtier, Félix Balazard, Rémy Dubois, Jeverson Moreira & Antoine Simon
Institut Curie, Paris, France
Armand Leopold, Maud Milder, Julien Guerin, Alain Livartowski & Guillaume Bataillon
Centre Léon Bérard, Lyon, France
Clément Joly, Julie Gervasoni, Thierry Durand & Pierre-Etienne Heudel
Institut Gustave Roussy, Villejuif, France
Damien Drubay & Magali Lacroix-Triki
Institut Universitaire du Cancer de Toulouse (IUCT) Oncopole, Toulouse, France
Camille Franchet
Peer review information
Nature Medicine thanks Roberto Salgado, Matthew Hanna and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Michael Basson and Javier Carmona, in collaboration with the Nature Medicine team.
Cite this article
Ogier du Terrail, J., Leopold, A., Joly, C. et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat Med (2023). https://doi.org/10.1038/s41591-022-02155-w
Originally published at https://www.nature.com on January 19, 2023.