This is a republication of an excerpt of the paper “Multi-omics microsampling for the profiling of lifestyle-associated changes in health”, with the title above. The post is preceded by an Executive Summary, by the Editor of the Site [Joaquim Cardoso]
institute for health transformation (inhealth)
Joaquim Cardoso MSc
Founder, CEO and Senior Advisor
January 23, 2023
EXECUTIVE SUMMARY
Current healthcare practices are reactive and use limited physiological and clinical information, often collected months or years apart.
- Moreover, the discovery and profiling of blood biomarkers in clinical and research settings are constrained by
(1) geographical barriers,
(2) the cost and inconvenience of in-clinic venepuncture,
(3) low sampling frequency and
(4) the low depth of molecular measurements.
Here we describe a strategy for the frequent capture and analysis of thousands of metabolites, lipids, cytokines and proteins in 10 μl of blood alongside physiological information from wearable sensors.
We show the advantages of such frequent and dense multi-omics microsampling in two applications:
- the assessment of the reactions to a complex mixture of dietary interventions, to discover individualized inflammatory and metabolic responses;
- and deep individualized profiling, to reveal large-scale molecular fluctuations as well as thousands of molecular relationships associated with intra-day physiological variations (in heart rate, for example) and with the levels of clinical biomarkers (specifically, glucose and cortisol) and of physical activity.
Combining wearables and multi-omics microsampling for frequent and scalable omics may facilitate dynamic health profiling and biomarker discovery.
Here we used two case studies to show the potential of multi-omics microsampling in precision medicine. Many other applications can be envisioned. Examples include:
- (1) Longitudinal biomarker discovery. The multi-omics microsampling is simple and unpainful compared with the traditional blood collection method and thus enables anyone to self-collect high-frequent and high-quality blood microsamples anywhere for longitudinal biomarker discovery.
- (2) Personalized health monitoring. People can collect blood samples at home without any help and then send the samples to the laboratory for data acquisition and analysis. If a notable abnormality is detected, the result is sent immediately to a physician. The physician would then be able to validate the results and respond quickly with an intervention.
- (3) Therapeutic drug monitoring. Patients could collect microsamples frequently and remotely to monitor the drug-related compounds or biomarkers in the blood at a known time, to guide dosage, and result in optimized therapy. In our study, all the microsamples were prepared and run together as a batch to avoid batch effects.
RELATED PUBLICATION

https://med.stanford.edu/news/all-news/2023/01/blood-drop-microsample-snyder.html
INFOGRAPHIC
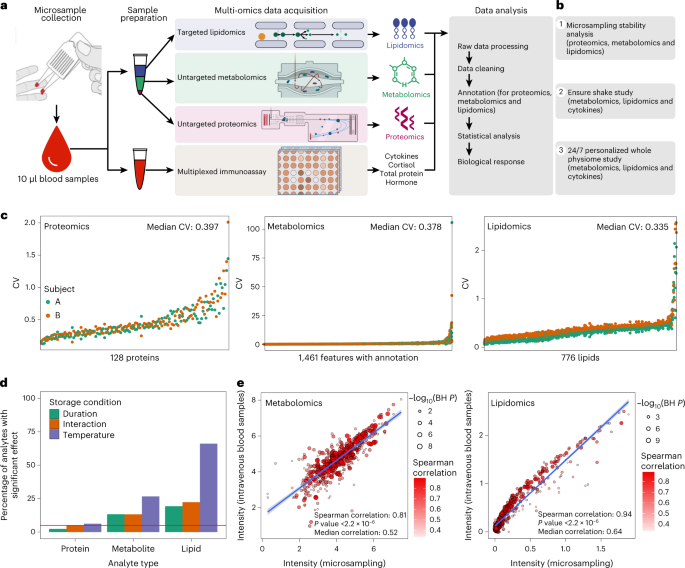
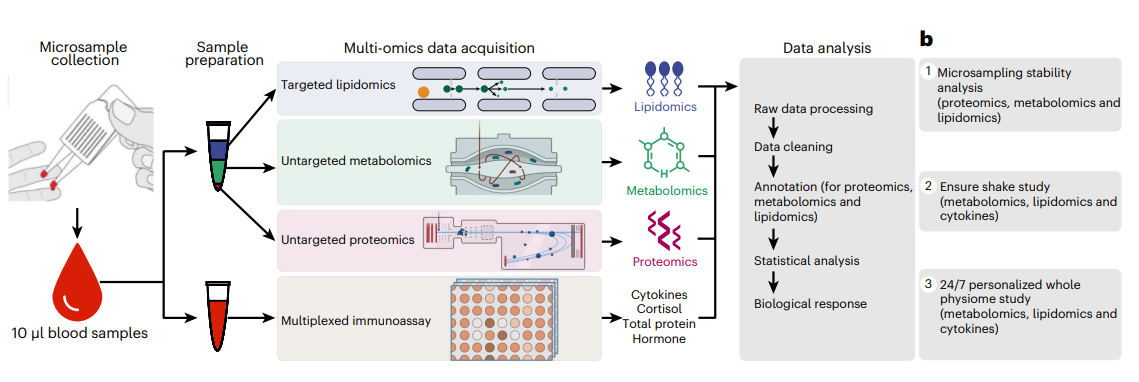
Fig. 1: Overview of the microsampling multi-omics workflow and stability analysis.
Case study 1: metabolic phenotyping responses to Ensure shake consumption [figures 2 and 3]
Fig. 2: The overview of Ensure shake study and molecular response to Ensure shake.
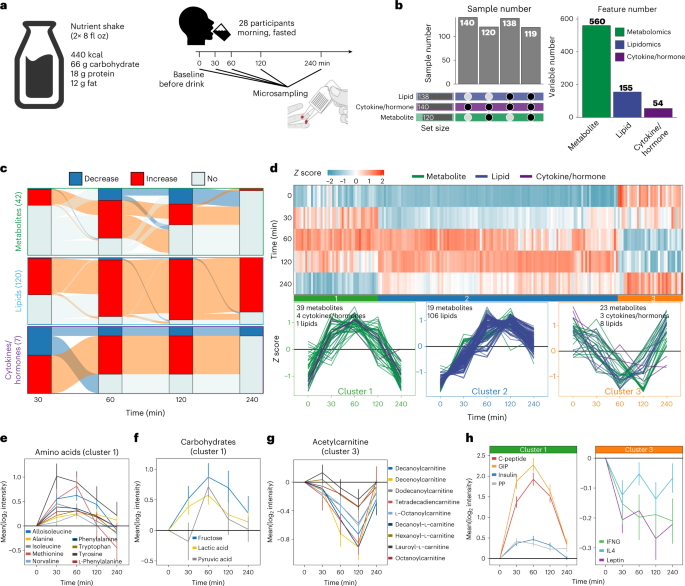
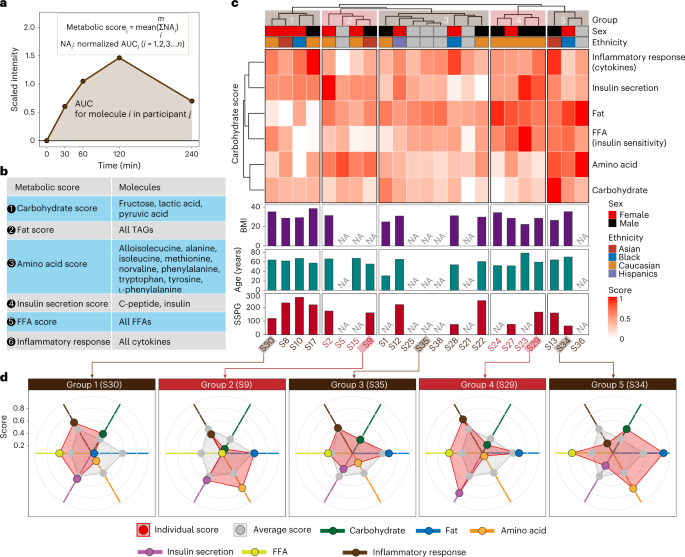
Fig. 3: Metabolic phenotyping based on the multi-omics response to the Ensure shake.
Case study 2: 24/7 personalized whole physiome profiling using wearable and multi-omics data [figures 4 , 5 and 6]
Fig. 4: Overview of the study design, sample collection and data acquisition for the 24/7 study.
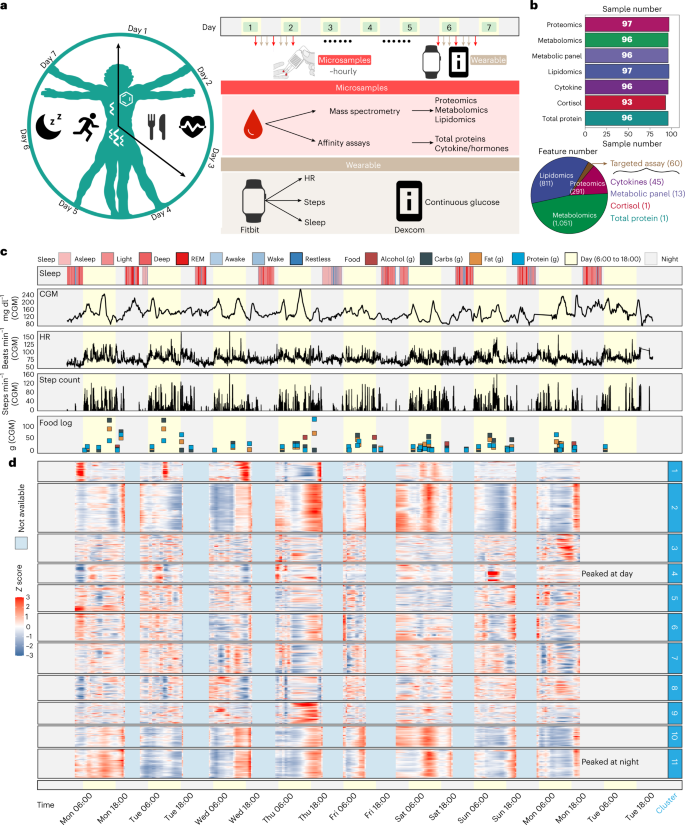
Fig. 5: Wearable and internal multi-omics data reflect the individual physiological status and circadian rhythm analysis of multi-omics data.
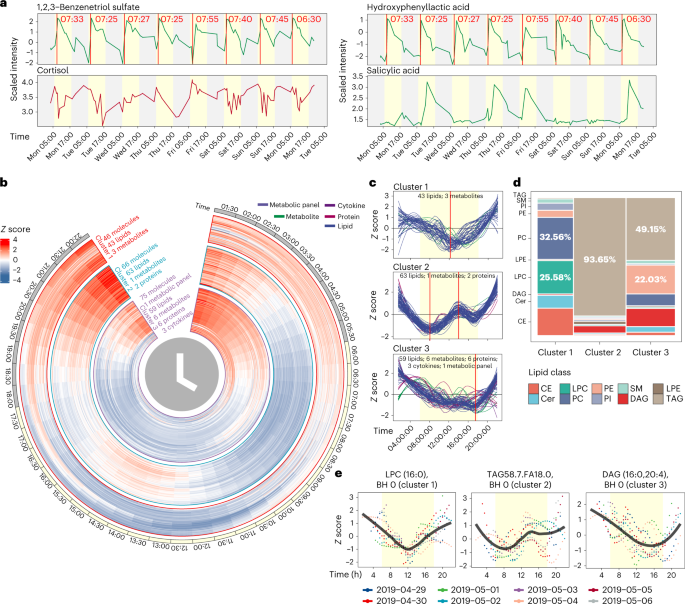
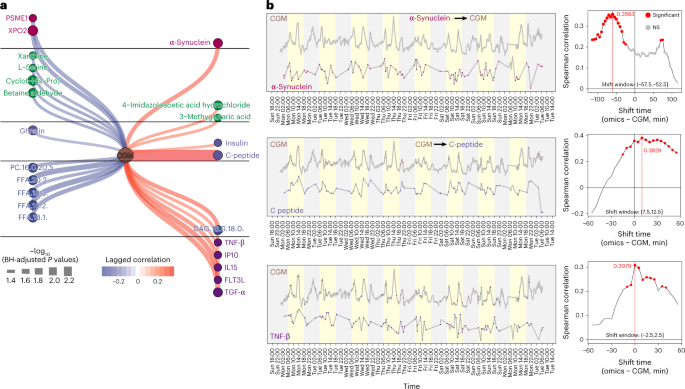
Fig. 6: CGM and internal molecule causal association network.
DEEP DIVE
Overview of the multi-omics microsampling approach
Nature Biomedical Engineering (2023)
Xiaotao Shen, Ryan Kellogg, Daniel J. Panyard, Nasim Bararpour, Kevin Erazo Castillo, Brittany Lee-McMullen, Alireza Delfarah, Jessalyn Ubellacker, Sara Ahadi, Yael Rosenberg-Hasson, Ariel Ganz, Kévin Contrepois, Basil Michael, Ian Simms, Chuchu Wang, Daniel Hornburg & Michael P. Snyder
Main
Multi-omics technologies enable the quantification of thousands of molecules and can provide new insights into the molecular landscape of health and disease1,2.
Despite major advances in omics technologies, the upstream sample collection and processing still requires travel to a clinic, access to a phlebotomist and physical and emotional discomfort.
These current sample-collection strategies do not meet the desired flexibility and non-invasiveness to conduct comprehensive longitudinal profiling independent of access to a clinic.
Furthermore, the high sample volume needed (often 10–50 ml of venous blood) prohibits frequent collections, which precludes high-resolution analysis of dynamic metabolic and biological processes that occur on the scale of minutes or hours.
Finally, high sample collection and processing costs can be prohibitive for performing large studies in remote environments.
Previous studies have investigated dried blood spot (DBS) sampling3,4,5,6 and volumetric absorptive microsampling (VAMS)7,8,9 for metabolite and protein analyses10.
In principle, DBS allows individuals to collect a blood drop sample at home and return the sample by mail at room temperature.
However, DBS sampling is often irreproducible since volumetric amounts can vary considerably, and, so far, the number of analytes analysed from DBS has generally been modest11.
In this Article, to circumvent these challenges, we devised a streamlined multi-omics profiling system that uses finger prick blood drop collection, minimizes pain and enables sampling frequencies on the timescale of minutes without needing clinic access.
Our method collects fixed 10 μl volumes and, following extraction, enables the simultaneous analysis of proteins, metabolites, lipids and targeted cytokines/hormones from a single sample enabling broad analyte profiling.
In two proof-of-principle studies, we first demonstrate the profiling of a dynamic response to ingestion of a mixed meal shake and discover high heterogeneity in individual metabolic and immune responses, and second, we perform high-resolution profiling of an individual over 1 week enabling the identification and quantification of thousands of molecular changes and associations across ‘omes’ at a personal level.
Our approach is scalable, enabling high-frequency molecular profiling for broad utility in research and clinical studies.
Results
See the original publication (this is an excerpt version)
Case studies
As a demonstration of the power of microsampling, we performed two case studies while participants were in their native environments.
The first was to examine the effect of drinking a complex mixture on metabolic profiles.
The second was to perform very dense ‘24/7’ profiling (98 microsamples) across a period of just longer than 7 days.
- Case study 1: metabolic phenotyping responses to Ensure shake consumption [figures 2 and 3]
- Case study 2: 24/7 personalized whole physiome profiling using wearable and multi-omics data [figures 4 , 5 and 6]
For details about the case studies, refer to the full version of the paper, please .

Discussion
Current healthcare practices are reactive and based on limited physiological and/or clinical information, often collected months or years apart.
In this study, we built a multi-omics microsampling approach that enables the measurement of thousands of metabolites, lipids, cytokines and proteins in frequently collected 10 μl blood samples.
We demonstrated that many of the molecules from the microsamples (VAMS) are stable and reliable.
In addition, most of the molecules from the microsampling are consistent with the classic blood sampling approach (Spearman correlation 0.8–0.9).
Compared with DBS, VAMS can achieve good analytical performance for targeted compound and protein analysis58,59.
However, for DBS, the haematocrit effect affects the resulting spot size, which can introduce variation in analysis.
As the microsampling approach is less invasive and can be used remotely and without specific training, it enables high-frequency blood sample collection (approximately hourly) in a native setting, which is difficult to perform using the classic blood sampling approach.
On the basis of the multi-omics microsampling workflow, we carried out two case studies to demonstrate the dense in situ samplings, and analytic capabilities to
(1) perform dynamic and individualized metabolic assessments after response to dietary (Ensure shake) intervention and
(2) reveal large-scale intra-day molecular fluctuations as well as thousands of molecular associations including those associated with intra-day variation in HR, glucose levels and activity.
It is worth noting that most analytes that we measure, particularly proteins, appear stable with regard to time, temperature and the combination of both.
We also note that, since we tested for significant effects of storage conditions with a relatively low sample size, we do not rule out additional effects that may not have been observed here due to power challenges, which was evident from a sensitivity regression analysis that analysed only one storage condition at a time (storage duration and storage temperature), and additional effects for storage duration were identified when the baseline samples were added to the analysis.
For those molecules that are not stable, they can either be discarded from the analyses or quantification can be ascertained from unique degradation products.
Alternatively, sample collection procedures could include rapid and cold shipping to minimize potential issues with less stable molecules. Indeed, we found that most samples can be collected and stored within 24 h, thus minimizing degradation.
Larger stability studies, especially in larger and more diverse populations, will help identify other potential issues.
Regardless, reliable measurements can be made for thousands of molecules, including those present at very low abundance (such as cytokines).

In summary, the presented methodology achieves fully remote, scalable, high-temporal-resolution omics and sensor monitoring.
It has the potential for large-scale comprehensive, dynamic molecular and digital biomarker discovery and monitoring as well as health profiling.
Here we used two case studies to show the potential of multi-omics microsampling in precision medicine. Many other applications can be envisioned.
Examples include:
(1) Longitudinal biomarker discovery. The multi-omics microsampling is simple and unpainful compared with the traditional blood collection method and thus enables anyone to self-collect high-frequent and high-quality blood microsamples anywhere for longitudinal biomarker discovery.
(2) Personalized health monitoring. People can collect blood samples at home without any help and then send the samples to the laboratory for data acquisition and analysis. If a notable abnormality is detected, the result is sent immediately to a physician. The physician would then be able to validate the results and respond quickly with an intervention.
(3) Therapeutic drug monitoring. Patients could collect microsamples frequently and remotely to monitor the drug-related compounds or biomarkers in the blood at a known time, to guide dosage, and result in optimized therapy. In our study, all the microsamples were prepared and run together as a batch to avoid batch effects.
In the future, the microsamples collected in 1 day could be prepared and run in 1 day after sample collection.
The users can receive their results within 2 days after sending their samples to the laboratory for analysis.
Additionally, developing a clinical diagnostic based on microsampling requires additional validation steps for accuracy, precision, matrix effects and so on, and the use of standards such as isotopically labelled reference molecules.
In addition, presently, only proteins, metabolites, lipids and cytokines were measured using our microsampling approach, but other types of molecules can be measured, such as DNA, epigenomes and RNA.
For the 24/7 study, as a pilot study, only one participant was recruited to demonstrate the power of following personalized responses.
Enlargement of the cohort size will enable the measurement of more generalized patterns but will also reveal new challenges in the processing and analysis of large numbers of samples.
Indeed, our simple studies generated 98 data points in a single individual.
Indeed, our simple studies generated 98 data points in a single individual.
The two pilot case studies (group study and individual study) were used to demonstrate the power and application of the approach.
The molecular signatures found in our study provide vast testable hypotheses that should be validated using analytical and experimental approaches.
We note that group analysis is usually performed to find the overall trend.
However, it can be potentially used to identify individual outliers who may have underlying conditions1.
When an individual profile differs greatly from the average, one needs to first check for sample mix-ups, systematic variation and batch effects.
Once normalized, data outlier detection can be further performed.
Individuals who fall outside the overall pattern can be investigated for underlying causes for their molecular shift (medical conditions, medications or lifestyle abnormalities).
In addition, the confounders (such as sex, age and body mass index (BMI)) must be controlled and adjusted to find the real and expected biological variation.
Similarly, we note that, when an individual profile differs greatly from the average, overview conclusions from the whole cohort may not extend to individuals 33,60.
For the personalized analysis, the conclusion from the individual may not extend to the group or other individuals60, which can be revealed using our approach.
Overall, we believe the multi-omics microsampling approach offers a promising opportunity to integrate with wearable data to improve precision healthcare.
Overall, we believe the multi-omics microsampling approach offers a promising opportunity to integrate with wearable data to improve precision healthcare.
Methods & Other sections
See the original publication
References
See the original publication
Originally published at https://www.nature.com on January 19, 2023.
About the authors & affiliations
Xiaotao Shen 1,2,5, Ryan Kellogg1,2,5, Daniel J. Panyard 1,2,5, Nasim Bararpour1,2,5, Kevin Erazo Castillo1,2, Brittany Lee-McMullen1,2, Alireza Delfarah1,2, Jessalyn Ubellacker1 , Sara Ahadi1,2, Yael Rosenberg-Hasson3 , Ariel Ganz 1,2, Kévin Contrepois1,2, Basil Michael1,2, Ian Simms1,2, Chuchu Wang 4 , Daniel Hornburg1,2 & Michael P. Snyder 1,
1 Department of Genetics, Stanford University School of Medicine, Stanford, CA, USA.
2 Stanford Center for Genomics and Personalized Medicine, Stanford, CA, USA.
3 Human Immune Monitoring Center, Microbiology and Immunology, Stanford University Medical Center, Stanford, CA, USA.
4 Howard Hughes Medical Institute, Stanford University, Stanford, CA, USA.












